PartSeg is python GUI and set of napari plugins for bio imaging analysis especially nucleus analysis,
Project description
PartSeg
PartSeg is a GUI and a library for segmentation algorithms. PartSeg also provide napari plugins for IO and labels measurement.
This application is designed to help biologist with segmentation based on threshold and connected components.
Tutorials
- Tutorial: Chromosome 1 (as gui) link
- Data for chromosome 1 tutorial link
- Tutorial: Different neuron types (as library) link
Installation
-
From binaries:
-
With pip:
- From pypi:
pip install PartSeg[all] - From repository:
pip install git+https://github.com/4DNucleome/PartSeg.git
- From pypi:
Installation troubleshooting information could be found in wiki: wiki. If this information does not solve problem you can open issue.
Running
If you downloaded binaries, run the PartSeg (or PartSeg.exe for Windows) file inside the PartSeg folder
If you installed from repository or from pip, you can run it with PartSeg command or python -m PartSeg.
First option does not work on Windows.
PartSeg export few commandline options:
--no_report- disable error reporting--no_dialog- disable error reporting and error dialog. Use only when running from terminal.segmentation_analysis- skip launcher and start analysis guisegmentation- skip launcher and start segmentation gui
napari plugin
PartSeg provides napari plugins for io to allow reading projects format in napari viewer.
Save Format
Saved projects are tar files compressed with gzip or bz2.
Metadata is saved in data.json file (in json format). Images/masks are saved as *.npy (numpy array format).
Interface
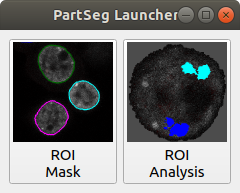
Launcher. Choose the program that you will launch:
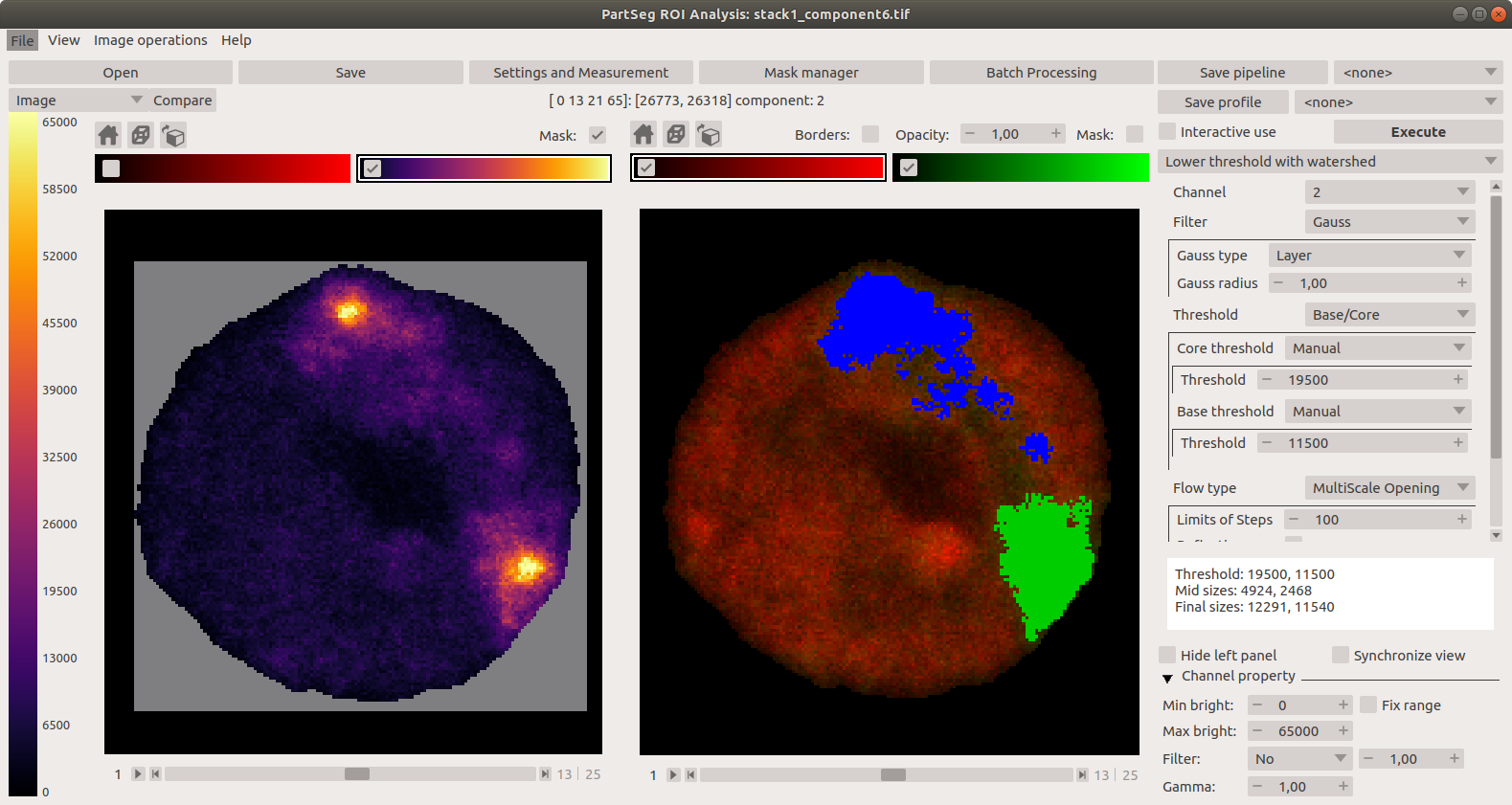
Main window of Segmentation Analysis:
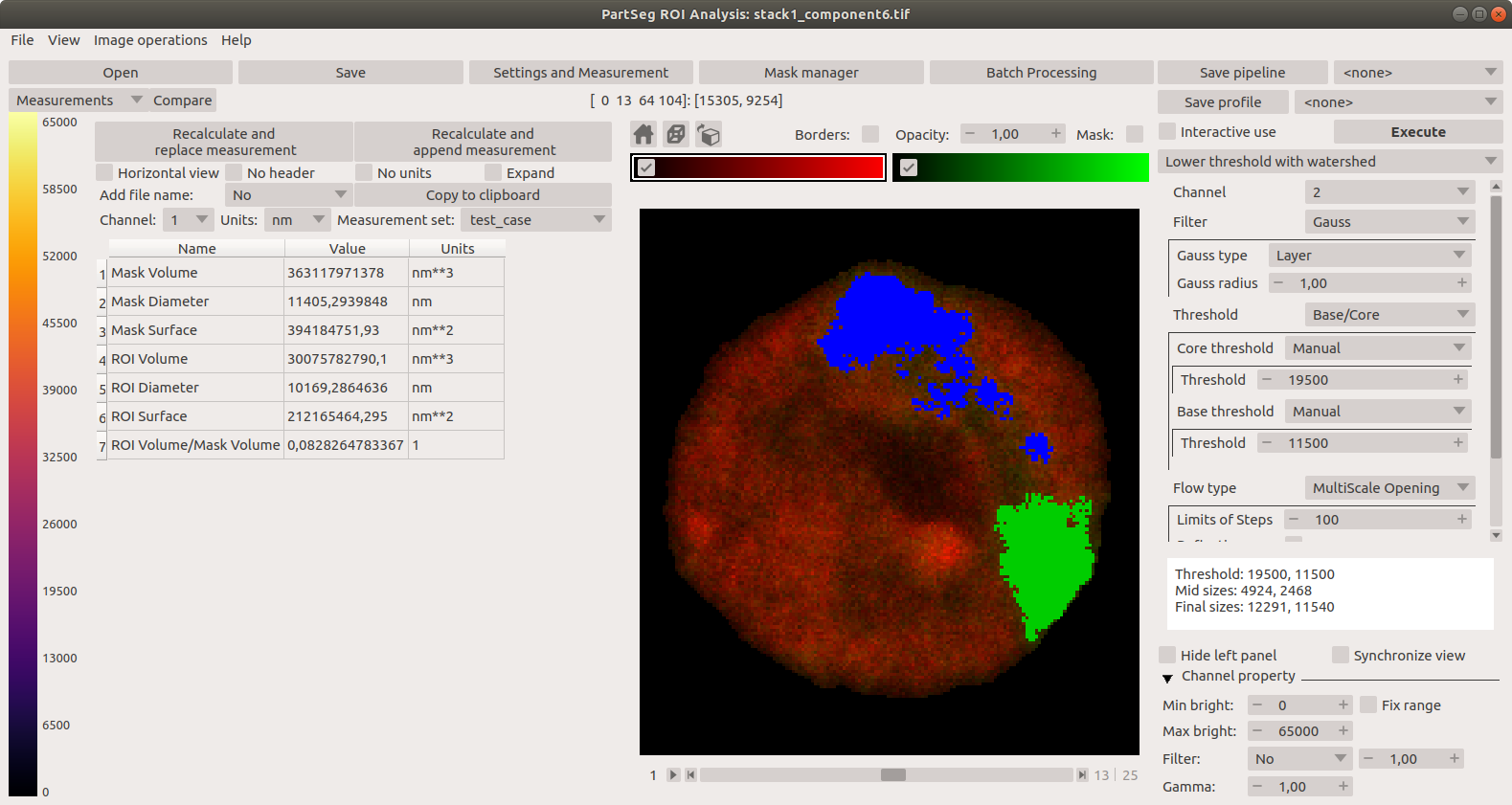
Main window of Segmentation Analysis with view on measurement result:
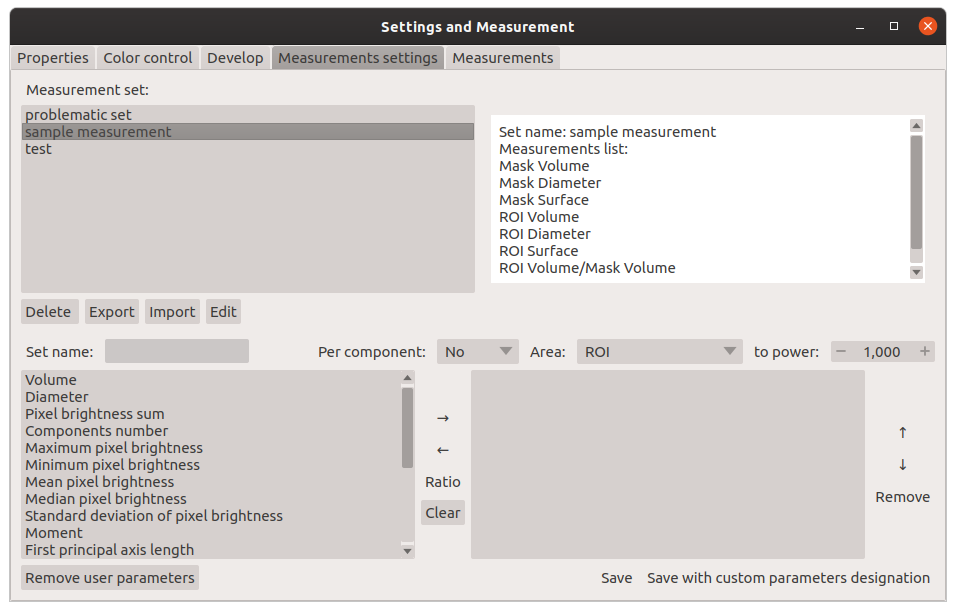
Window for creating a set of measurements:
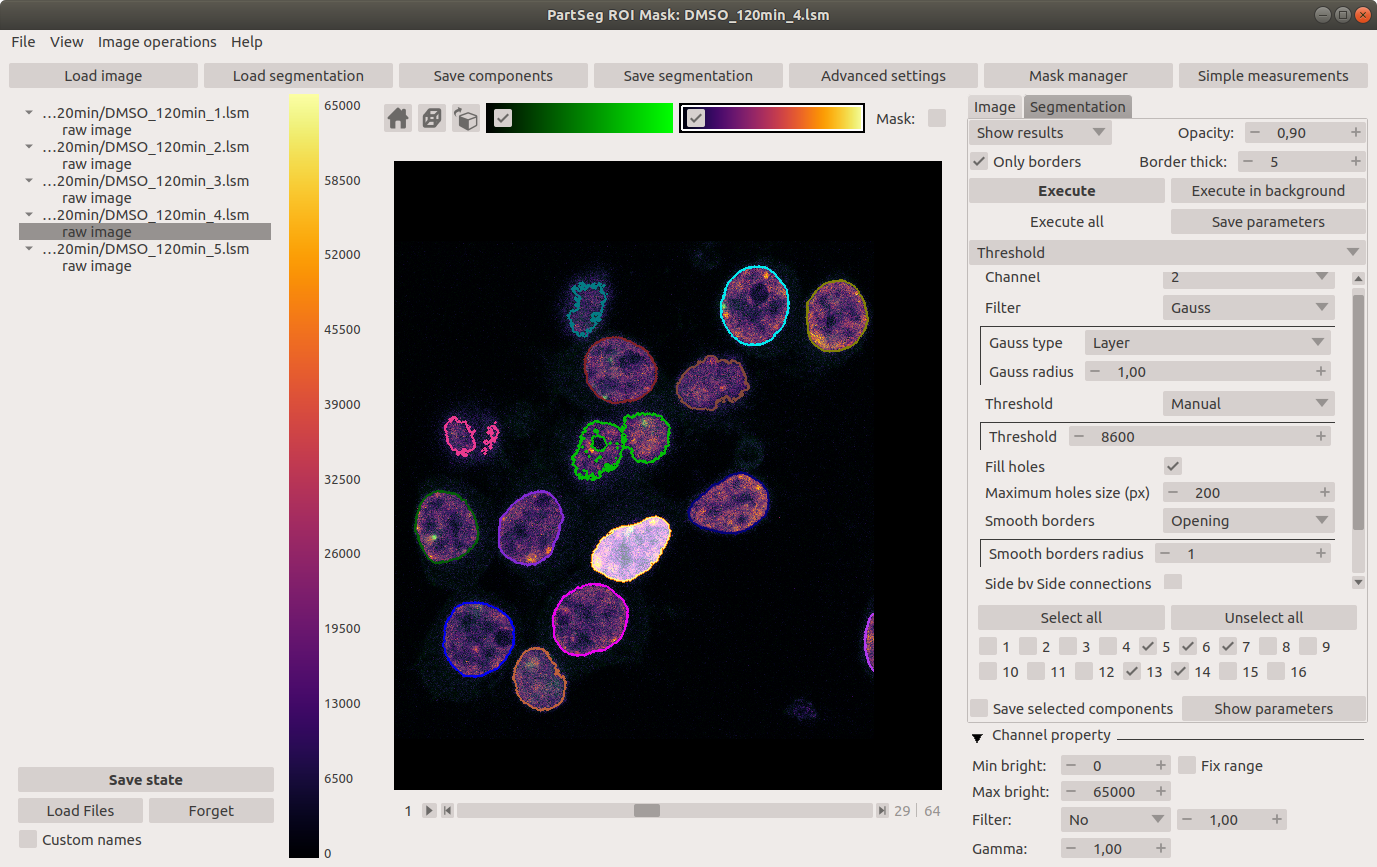
Main window of Mask Segmentation:
Laboratory
Laboratory of Functional and Structural Genomics http://4dnucleome.cent.uw.edu.pl/
Cite as
Bokota, G., Sroka, J., Basu, S. et al. PartSeg: a tool for quantitative feature extraction from 3D microscopy images for dummies. BMC Bioinformatics 22, 72 (2021). https://doi.org/10.1186/s12859-021-03984-1
Changelog
0.13.14
Bug Fixes
- ROI alternative representation (#471)
- Change additive to translucent in rendering ROI and Mask (#472)
Features
- Add morphological watershed segmentation (#469)
- Add Bilateral image filter (#470)
0.13.13
Bug Fixes
- Fix bugs in the generation process of the changelog for release. (#428)
- Restoring ROI on home button click in compare viewer (#443)
- Fix Measurement name prefix in bundled PartSeg. (#458)
- Napari widgets registration in pyinstaller bundle (#465)
- Hide points button if no points are loaded, hide Mask checkbox if no mask is set (#463)
- Replace Label data instead of adding/removing layers - fix blending layers (#464)
Features
- Add threshold information in layer annotation in the Multiple Otsu ROI extraction method (#430)
- Add option to select rendering method for ROI (#431)
- Add callback mechanism to ProfileDict, live update of ROI render parameters (#432)
- Move the info bar on the bottom of the viewer (#442)
- Add options to load recent files in multiple files widget (#444)
- Add ROI annotations as properties to napari labels layer created by ROI Extraction widgets (#445)
- Add signals to ProfileDict, remove redundant synchronization mechanisms (#449)
- Allow ignoring updates for 21 days (#453)
- Save all components if no components selected in mask segmentation (#456)
- Add modal dialog for search ROI components (#459)
- Add full measurement support as napari widget (#460)
- Add search labels as napari widget (#467)
Refactor
- Export common code for load/save dialog to one place (#437)
- Change most of call QFileDialog to more generic code (#440)
Testing
- Add test for
PartSeg.common_backendmodule (#433)
0.13.12
Bug Fixes
- Importing the previous version of settings (#406)
- Cutting without masking data (#407)
- Save in subdirectory in batch plan (#414)
- Loading plugins for batch processing (#423)
Features
- Add randomization option for correlation calculation (#421)
- Add Imagej TIFF writter for image. (#405)
- Mask create widget for napari (#395)
- In napari roi extraction method show information from roi extraction method (#408)
- Add
*[0-9].tifbutton in batch processing window (#412) - Better label representation in 3d view (#418)
Refactor
- Use Font Awesome instead of custom symbols (#424)
0.13.11
Bug Fixes
- Adding mask in Prepare Plan for batch (#383)
- Set proper completion mode in SearchComboBox (#384)
- Showing warnings on the error with ROI load (#385)
Features
- Add CellFromNucleusFlow "Cell from nucleus flow" cell segmentation method (#367)
- When cutting components in PartSeg ROI mask allow not masking outer data (#379)
- Theme selection in GUI (#381)
- Allow return points from ROI extraction algorithm (#382)
- Add measurement to get ROI annotation by name. (#386)
- PartSeg ROI extraction algorithms as napari plugins (#387)
- Add Pearson, Mander's, Intensity, Spearman colocalization measurements (#392)
- Separate standalone napari settings from PartSeg embedded napari settings (#397)
Performance
- Use faster calc bound function (#375)
Refactor
- Remove CustomApplication (#389)
0.13.10
- change tiff save backend to ome-tiff
- add
DistanceROIROIandROINeighbourhoodROImeasurements
0.13.9
- annotation show bugfix
0.13.8
- napari deprecation fixes
- speedup simple measurement
- bundle plugins initial support
0.13.7
- add measurements widget for napari
- fix bug in pipeline usage
0.13.6
- Hotfix release
- Prepare for a new napari version
0.13.5
- Small fixes for error reporting
- Fix mask segmentation
0.13.4
- Bugfix for outdated profile/pipeline preview
0.13.3
- Fix saving roi_info in multiple files and history
0.13.2
- Fix showing label in select label tab
0.13.1
- Add Haralick measurements
- Add obsep file support
0.13.0
- Add possibility of custom input widgets for algorithms
- Switch to napari Colormaps instead of custom one
- Add points visualization
- Synchronization widget for builtin (View menu) napari viewer
- Drop Python 3.6
0.12.7
- Fixes for napari 0.4.6
0.12.6
- Fix prev_mask_get
- Fix cache mechanism on mask change
- Update PyInstaller build
0.12.5
- Fix bug in pipeline execute
0.12.4
- Fix ROI Mask windows related build (signal not properly connected)
0.12.3
- Fix ROI Mask
0.12.2
- Fix windows bundle
0.12.1
- History of last opened files
- Add ROI annotation and ROI alternatives
- Minor bugfix
0.12.0
- Toggle multiple files widget in View menu
- Toggle Left panel in ROI Analysis in View Menu
- Rename Mask Segmentation to ROI Mask
- Add documentation for interface
- Add Batch processing tutorial
- Add information about errors to batch processing output file
- Load image from the batch prepare window
- Add search option in part of list and combo boxes
- Add drag and drop mechanism to load list of files to batch window.
0.11.5
- add side view to viewer
- fix horizontal view for Measurements result table
0.11.4
- bump to napari 0.3.8 in bundle
- fix bug with not presented segmentation loaded from project
- add frame (1 pix) to image cat from base one based on segmentation
- pin to Qt version to 5.14
0.11.3
- prepare for napari 0.3.7
- split napari io plugin on multiple part
- better reporting for numpy array via sentry
- fix setting color for mask marking
0.11.2
- Speedup image set in viewer using async calls
- Fix bug in long name of sheet with parameters
0.11.1
- Add screenshot option in View menu
- Add Voxels measurements
0.11.0
- Make sprawl algorithm name shorter
- Unify capitalisation of measurement names
- Add simple measurements to mask segmentation
- Use napari as viewer
- Add possibility to preview additional output of algorithms (In View menu)
- Update names of available Algorithm and Measurement to be more descriptive.
0.10.8
- fix synchronisation between viewers in Segmentation Analysis
- fix batch crash on error during batch run, add information about file on which calculation fails
- add changelog preview in Help > About
0.10.7
- in measurements, on empty list of components mean will return 0
0.10.6
- fix border rim preview
- fix problem with size of image preview
- zoom with scroll and moving if rectangle zoom is not marked
0.10.5
- make PartSeg PEP517 compatible.
- fix multiple files widget on Windows (path normalisation)
0.10.4
- fix slow zoom
0.10.3
- deterministic order of elements in batch processing.
0.10.2
- bugfixes
0.10.1
- bugfixes
0.10.0
- Add creating custom label coloring.
- Change execs interpreter to python 3.7.
- Add masking operation in Segmentation Mask.
- Change license to BSD.
- Allow select root type in batch processing.
- Add median filter in preview.
0.9.7
- fix bug in compare mask
0.9.6
- fix bug in loading project with mask
- upgrade PyInstaller version (bug GHSA-7fcj-pq9j-wh2r)
0.9.5
- fix bug in loading project in "Segmentation analysis"
0.9.4
- read mask segmentation projects
- choose source type in batch
- add initial support to OIF and CZI file format
- extract utils to PartSegCore module
- add automated tests of example notebook
- reversed mask
- load segmentation parameters in mask segmentation
- allow use sprawl in segmentation tool
- add radial split of mask for measurement
- add all measurement results in batch, per component sheet
0.9.3
- start automated build documentation
- change color map backend and allow for user to create custom color map.
- segmentation compare
- update test engines
- support of PySide2
0.9.2.3
- refactor code to make easier create plugin for mask segmentation
- create class base updater for update outdated algorithm description
- fix save functions
- fix different bugs
0.9.2.2
- extract static data to separated package
- update marker of fix range and add mark of gauss in channel control
0.9.2.1
- add VoteSmooth and add choosing of smooth algorithm
0.9.2
-
add pypi base check for update
-
remove resetting image state when change state in same image
-
in stack segmentation add options to picking components from segmentation's
-
in mask segmentation add:
- preview of segmentation parameters per component,
- save segmentation parameters in save file
- new implementation of batch mode.
0.9.1
-
Add multiple files widget
-
Add Calculating distances between segmented object and mask
-
Batch processing plan fixes:
- Fix adding pipelines to plan
- Redesign mask widget
-
modify measurement backend to allow calculate multi channel measurements.
0.9
Begin of changelog
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file PartSeg-0.13.14.tar.gz.
File metadata
- Download URL: PartSeg-0.13.14.tar.gz
- Upload date:
- Size: 390.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.8.1 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.9.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 4b23fe94e3ff54764befdaa5b24f01eb256d84ba33a44815148325bc97d070ec |
|
| MD5 | 2623dafe58bf136e0c8ba4b75996f7cd |
|
| BLAKE2b-256 | 4649ed4e3939a4c6d337835c33375645ead450898aacac6d4f6bb3e01a37e7b6 |
File details
Details for the file PartSeg-0.13.14-py3-none-any.whl.
File metadata
- Download URL: PartSeg-0.13.14-py3-none-any.whl
- Upload date:
- Size: 330.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.8.1 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.9.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 67213a91b2aa6ce0be069b591e6f160ac6b7d5986852bf2a535afbbfd6268dec |
|
| MD5 | 66990fe801ec9392a2ca068ad3cdde64 |
|
| BLAKE2b-256 | 3567f774f60fcc622beedd35efa278e98790c9d35215b319f4792aa16cf4d728 |