Operations and utilities for Genomic Interval Dataframes.
Project description
Bioframe: Operations on Genomic Interval Dataframes

Bioframe enables flexible and scalable operations on genomic interval dataframes in Python.
Bioframe is built directly on top of Pandas. Bioframe provides:
- A variety of genomic interval operations that work directly on dataframes.
- Operations for special classes of genomic intervals, including chromosome arms and fixed-size bins.
- Conveniences for diverse tabular genomic data formats and loading genome assembly summary information.
Read the docs, including the guide, as well as the bioframe preprint for more information.
Bioframe is an Affiliated Project of NumFOCUS.
Installation
Bioframe is available on PyPI and bioconda:
pip install bioframe
Contributing
Interested in contributing to bioframe? That's great! To get started, check out the contributing guide. Discussions about the project roadmap take place on the Open2C Slack and regular developer meetings scheduled there. Anyone can join and participate!
Interval operations
Key genomic interval operations in bioframe include:
overlap: Find pairs of overlapping genomic intervals between two dataframes.closest: For every interval in a dataframe, find the closest intervals in a second dataframe.cluster: Group overlapping intervals in a dataframe into clusters.complement: Find genomic intervals that are not covered by any interval from a dataframe.
Bioframe additionally has functions that are frequently used for genomic interval operations and can be expressed as combinations of these core operations and dataframe operations, including: coverage, expand, merge, select, and subtract.
To overlap two dataframes, call:
import bioframe as bf
bf.overlap(df1, df2)
For these two input dataframes, with intervals all on the same chromosome:
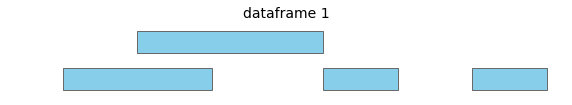

overlap will return the following interval pairs as overlaps:
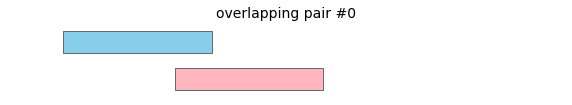
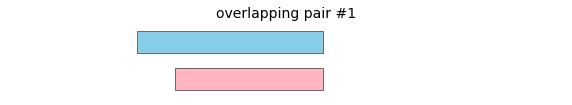
To merge all overlapping intervals in a dataframe, call:
import bioframe as bf
bf.merge(df1)
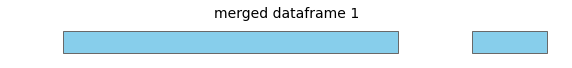
For this input dataframe, with intervals all on the same chromosome:

merge will return a new dataframe with these merged intervals:
See the guide for visualizations of other interval operations in bioframe.
File I/O
Bioframe includes utilities for reading genomic file formats into dataframes and vice versa. One handy function is read_table which mirrors pandas’s read_csv/read_table but provides a schema argument to populate column names for common tabular file formats.
jaspar_url = 'http://expdata.cmmt.ubc.ca/JASPAR/downloads/UCSC_tracks/2022/hg38/MA0139.1.tsv.gz'
ctcf_motif_calls = bioframe.read_table(jaspar_url, schema='jaspar', skiprows=1)
Tutorials
See this jupyter notebook for an example of how to assign TF motifs to ChIP-seq peaks using bioframe.
Citing
If you use bioframe in your work, please cite:
@article{bioframe_2022,
author = {Open2C and Abdennur, Nezar and Fudenberg, Geoffrey and Flyamer, Ilya and Galitsyna, Aleksandra and Goloborodko, Anton and Imakaev, Maxim and Venev, Sergey},
doi = {10.1101/2022.02.16.480748},
journal = {bioRxiv},
title = {{Bioframe: Operations on Genomic Intervals in Pandas Dataframes}},
year = {2022}
}
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file bioframe-0.6.2.tar.gz.
File metadata
- Download URL: bioframe-0.6.2.tar.gz
- Upload date:
- Size: 952.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/5.0.0 CPython/3.12.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | b9519206115ae435c18ec57f9ae49768851202684949caa7d183ac76f812e5c7 |
|
| MD5 | 53a33e4617845de927a07a92b3fd1269 |
|
| BLAKE2b-256 | 1d0bb20e27302f244ed0cd4b65e9637ed12d18b00ca73bcf7c4ea93d443797a2 |
File details
Details for the file bioframe-0.6.2-py2.py3-none-any.whl.
File metadata
- Download URL: bioframe-0.6.2-py2.py3-none-any.whl
- Upload date:
- Size: 144.0 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? Yes
- Uploaded via: twine/5.0.0 CPython/3.12.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 6d44a44a8e8e8953589fb8d61921f5f386ed1b485d675c0bcc6dbf5d453cbcdc |
|
| MD5 | d93b70235d09964c4969c1d34a7ecd55 |
|
| BLAKE2b-256 | b411f9b067eb34067a3904a1c6d6d2ec5b73dd1c1b03f8d21c87e3c5afee5df4 |



















