The Pythonic Blue Brain simulator access
Project description
BlueCelluLab
Latest Release |
|
Documentation |
|
License |
|
Build Status |
|
Coverage |
|
Gitter |
|
Citation |
The Blue Brain Cellular Laboratory is designed for simulations and experiments on individual cells or groups of cells. Suitable use cases for BlueCelluLab include:
Scripting and statistical analysis for single cells or cell pairs.
Lightweight, detailed reporting on specific state variables after simulation.
Developing synaptic plasticity rules.
Validating dynamics of synaptic properties.
Automating in-silico whole-cell patching experiments.
Debugging, both scientifically and computationally.
Citation
When you use this BlueCelluLab software for your research, we ask you to cite the following reference(this includes poster presentations):
@software{bluecellulab_zenodo,
author = {Van Geit, Werner and Tuncel, Anil and Gevaert, Mike and Torben-Nielsen, Benjamin and Muller, Eilif},
title = {BlueCelluLab},
month = jul,
year = 2023,
publisher = {Zenodo},
doi = {10.5281/zenodo.8113483},
url = {https://doi.org/10.5281/zenodo.8113483}
}Support
We are providing support on Gitter. We suggest you create tickets on the Github issue tracker in case you encounter problems while using the software or if you have some suggestions.
Main dependencies
Installation
BlueCelluLab can be pip installed with the following command:
pip install bluecellulabQuick Start
The following example shows how to create a cell, add a stimulus and run a simulation:
from bluecellulab.cell import create_ball_stick
from bluecellulab import Simulation
cell = create_ball_stick()
sim = Simulation()
sim.add_cell(cell)
stimulus = cell.add_step(start_time=5.0, stop_time=20.0, level=0.5)
sim.run(25, cvode=False)
time, voltage = cell.get_time(), cell.get_soma_voltage()
# plotting time and voltage ...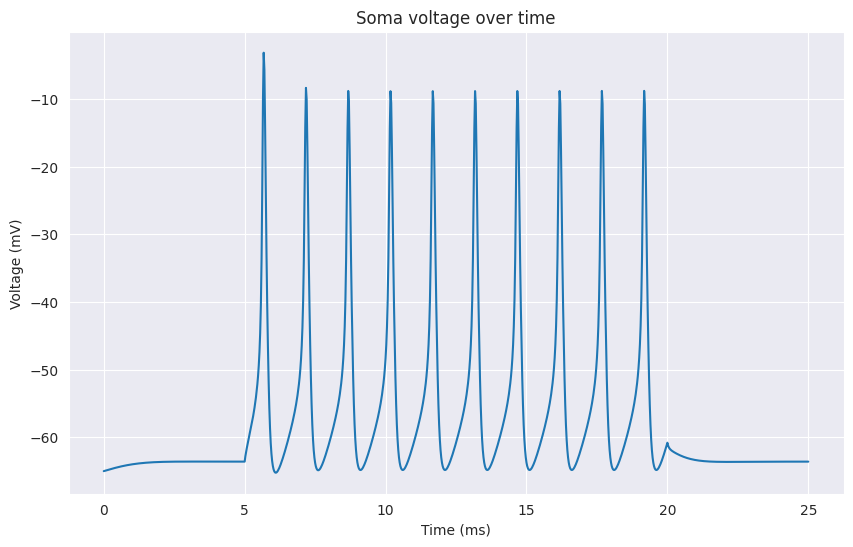
Tutorial
A more detailed explanation on how to use BlueCelluLab, as well as other examples can be found on the examples page.
API Documentation
The API documentation can be found on ReadTheDocs.
Funding & Acknowledgements
The development and maintenance of this code is supported by funding to the Blue Brain Project, a research center of the École polytechnique fédérale de Lausanne (EPFL), from the Swiss government’s ETH Board of the Swiss Federal Institutes of Technology.
Copyright
Copyright (c) 2023 Blue Brain Project/EPFL
This work is licensed under Apache 2.0
For MOD files for which the original source is available on ModelDB, any specific licenses on mentioned on ModelDB, or the generic License of ModelDB apply.
The licenses of the morphology files used in this repository are available on: https://zenodo.org/record/5909613
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file bluecellulab-1.7.2.tar.gz.
File metadata
- Download URL: bluecellulab-1.7.2.tar.gz
- Upload date:
- Size: 15.6 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.11.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | c4b54e6d80fe4b66a8d70ddd6061d4636f76cbdf0c0c8fe4a50ce269013ee111 |
|
| MD5 | ad30b341f3d2f0c8ef51668cdff62bf7 |
|
| BLAKE2b-256 | 485a5bd88c5589e1e9cfc27fe5de51125ad61326a00116eb5a17bf516a42b5a8 |
File details
Details for the file bluecellulab-1.7.2-py3-none-any.whl.
File metadata
- Download URL: bluecellulab-1.7.2-py3-none-any.whl
- Upload date:
- Size: 107.3 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.11.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | c8575a265977a10ddd63498b6a27d2b0b37d152b36a9e5f1aa01787f8a3f201a |
|
| MD5 | 1fc6b9e876807fb8171fa5ffa4e3a070 |
|
| BLAKE2b-256 | eae1dcaa13748d2944bddadaa43f47c2631626e9584e9e41d7c4db6783082679 |





















