Blue Brain Python Electrical Modeling Pipeline
Project description
BluePyEModel: Blue Brain Python Electrical Modeling Pipeline
Latest Release |
|
Documentation |
|
License |
|
Build Status |
|
Coverage |
|
Citation |
Introduction
The Blue Brain Python Electrical Modeling Pipeline (BluePyEModel) is a Python package facilitating the configuration and execution of electrical neuron model (e-model) building tasks. It covers tasks such as extraction of electrical features from electrophysiology data, e-model parameters optimisation and model validation. As such, it builds on top of eFEL, BluePyEfe and BluePyOpt.
For a general overview and example of electrical model building, please refer to the paper: A universal workflow for creation, validation and generalization of detailed neuronal models.
Note that this package only covers e-model building based on patch-clamp data and that it relies solely on the NEURON simulator.
Citation
When you use the BluePyEModel software or method for your research, we ask you to cite the following publication (this includes poster presentations):
@software{bluepyemodel_zenodo,
author = {Damart, Tanguy and Jaquier, Aurélien and Arnaudon, Alexis and Mandge, Darshan and Van Geit, Werner and Kilic, Ilkan},
title = {BluePyEModel},
month = aug,
year = 2023,
publisher = {Zenodo},
doi = {8283490},
url = {https://doi.org/10.5281/zenodo.8283490}
}Installation
BluePyEModel can be pip installed with the following command:
pip install bluepyemodel[all]If you do not wish to install all dependencies, specific dependencies can be selected by indicating which ones to install between brackets in place of ‘all’ (If you want multiple dependencies, they have to be separated by commas). The available dependencies are:
luigi
all
To get started with the E-Model building pipeline
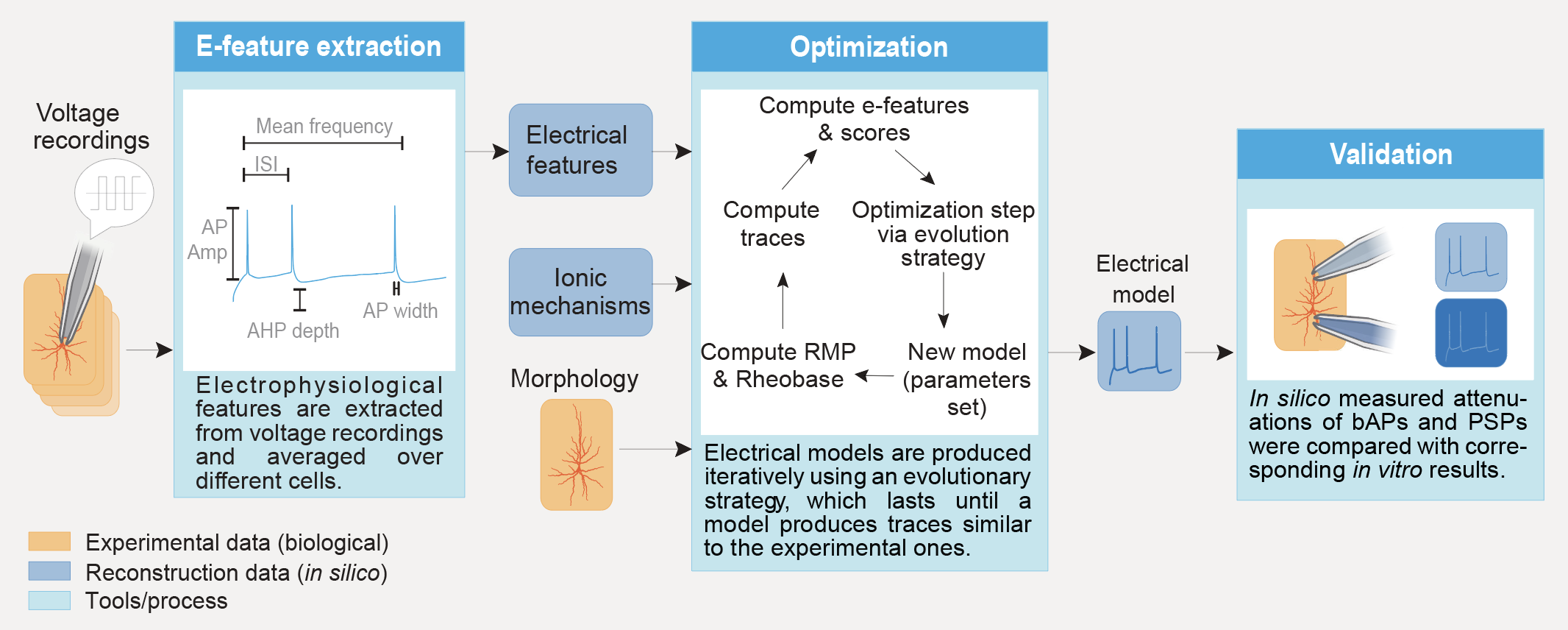
This section presents a general picture of the pipeline. For a detailed picture and how to use it, please refer to the example directory and its README.
The pipeline is divided in 6 steps:
extraction: extracts e-features from ephys recordings and averages the results e-feature values along the requested targets.
optimisation: builds a NEURON cell model and optimises its parameters using as targets the efeatures computed during e-feature extraction.
storage of the model: reads the results of the extraction and stores the models (best set of parameters) in a local json file.
validation: reads the models and runs the optimisation protocols and/or validation protocols on them. The e-feature scores obtained on these protocols are then passed to a validation function that decides if the model is good enough.
plotting: reads the models and runs the optimisation protocols and/or validation protocols on them. Then, plots the resulting traces along the e-feature scores and parameter distributions.
exporting: read the parameter of the best models and export them in files that can be used either in NEURON or for circuit building.
These six steps are to be run in order as for example validation cannot be run if no models have been stored. Steps “validation”, “plotting” and “exporting” are optional. Step “extraction” can also be optional in the case where the file containing the protocols and optimisation targets is created by hand or if it is obtained from an older project.
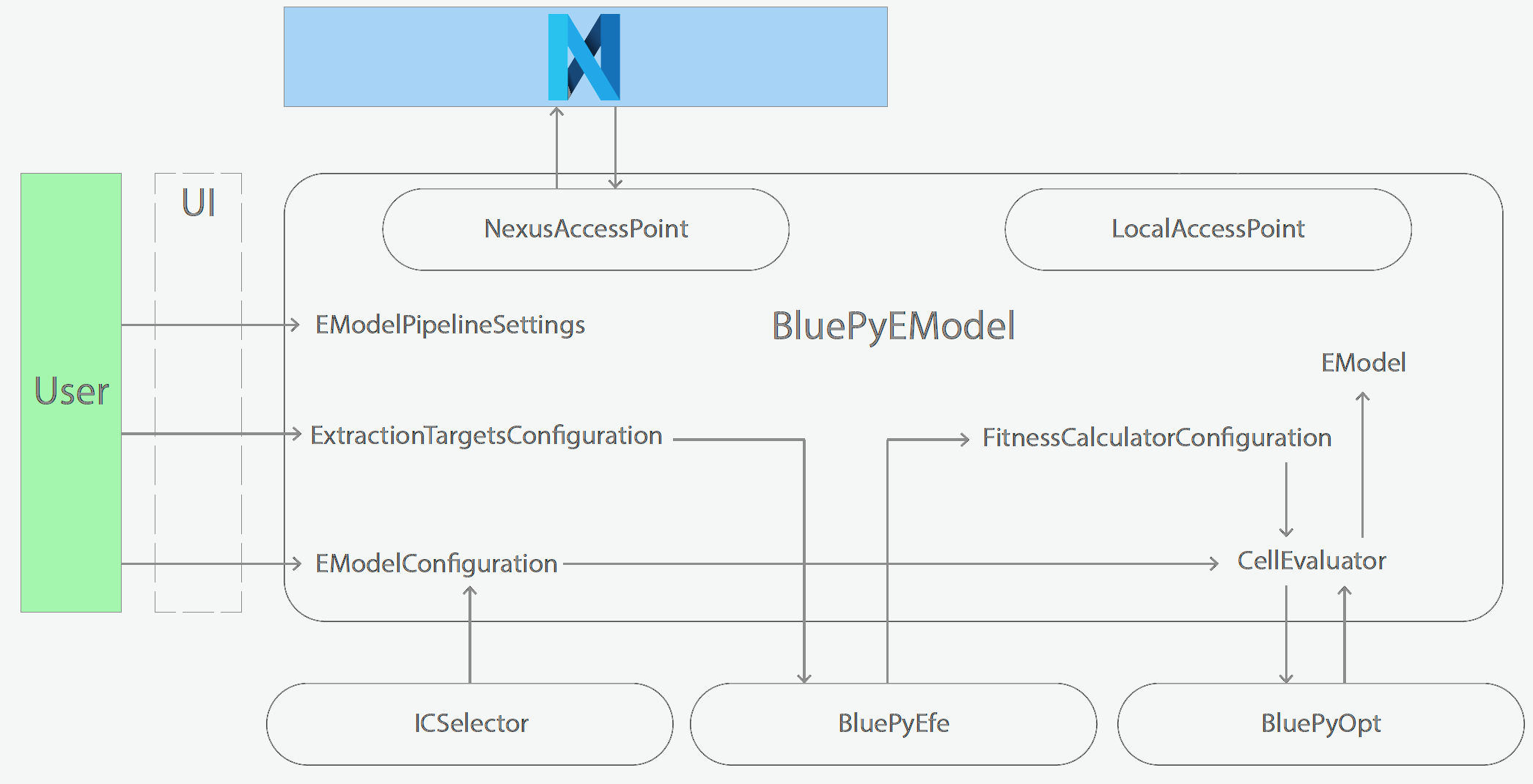
Schematics of BluePyEModel classes
Acknowledgment
This work was supported by funding to the Blue Brain Project, a research center of the École polytechnique fédérale de Lausanne (EPFL), from the Swiss government’s ETH Board of the Swiss Federal Institutes of Technology. This work has been partially funded by the European Union Seventh Framework Program (FP7/20072013) under grant agreement no. 604102 (HBP), and by the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreements No. 720270 (Human Brain Project SGA1) and No. 785907 (Human Brain Project SGA2) and by the EBRAINS research infrastructure, funded from the European Union’s Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 945539 (Human Brain Project SGA3).
Copyright
Copyright (c) 2023 Blue Brain Project/EPFL
This work is licensed under Apache 2.0
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file bluepyemodel-0.0.120.tar.gz.
File metadata
- Download URL: bluepyemodel-0.0.120.tar.gz
- Upload date:
- Size: 10.2 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.12.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | af35b3420887e1c76868b33909a860d4dc3914b6337f8029336d9b8b3f280d97 |
|
| MD5 | f5858c52806d198e81e58ddca70e9ad0 |
|
| BLAKE2b-256 | 0fe09aec0a2fdd4427add55af1b9869b5fd228432bc51da1bea37623dd6b5b97 |
File details
Details for the file bluepyemodel-0.0.120-py3-none-any.whl.
File metadata
- Download URL: bluepyemodel-0.0.120-py3-none-any.whl
- Upload date:
- Size: 9.6 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.12.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 6937d856853008431ac1e400980ab64f7443544ccbc4149db372e5ed3cd422a7 |
|
| MD5 | fd356e943765878bba91b5625e46ae75 |
|
| BLAKE2b-256 | 4dd731651b5b8e8ef812d1e2de195b7eca007bf2d4271a4b34a2f9b37c842934 |