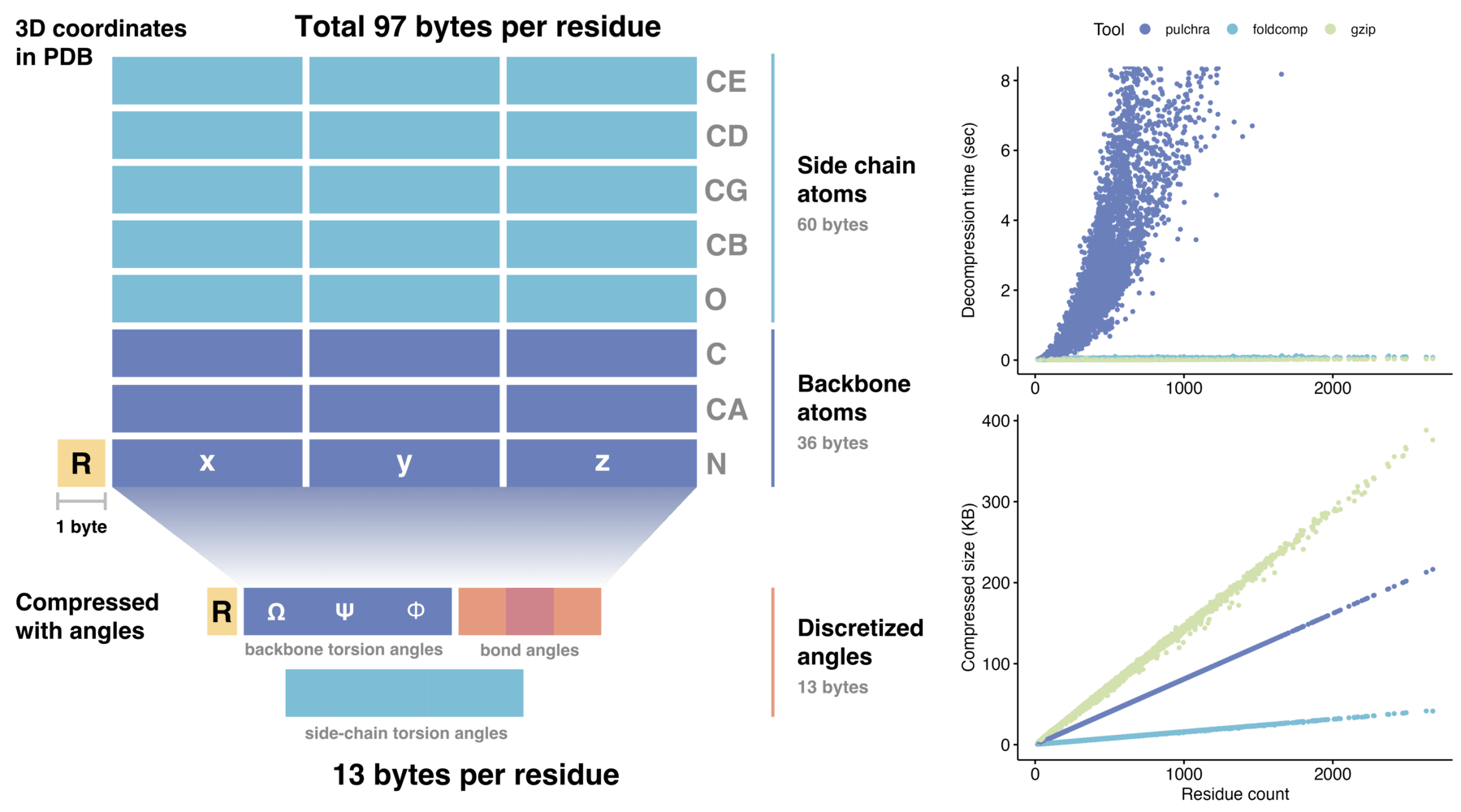
Foldcomp compresses protein structures with torsion angles effectively. It compresses the backbone atoms to 8 bytes and the side chain to additionally 4-5 byes per residue, an averaged-sized protein of 350 residues requires ~4.2kb. Foldcomp is a C++ library with Python bindings.
Project description
Foldcomp

Foldcomp efficient compressed format stores protein structures requiring only 13 bytes per residue, which reduces the required storage space by an order of magnitude compared to saving 3D coordinates directly. We achieve this reduction by encoding the torsion angles of the backbone as well as the side-chain angles in a compact binary file format (FCZ).
Foldcomp currently only supports compression of single chain PDB files
Publications
Usage
Installing Foldcomp
# Install Foldcomp Python package
pip install foldcomp
# Download static binaries for Linux
wget https://mmseqs.com/foldcomp/foldcomp-linux-x86_64.tar.gz
# Download static binaries for Linux (ARM64)
wget https://mmseqs.com/foldcomp/foldcomp-linux-arm64.tar.gz
# Download binary for macOS
wget https://mmseqs.com/foldcomp/foldcomp-macos-universal.tar.gz
# Download binary for Windows (x64)
wget https://mmseqs.com/foldcomp/foldcomp-windows-x64.zip
Executable
# Compression
foldcomp compress <pdb|cif> [<fcz>]
foldcomp compress [-t number] <dir|tar(.gz)> [<dir|tar|db>]
# Decompression
foldcomp decompress <fcz|tar> [<pdb>]
foldcomp decompress [-t number] <dir|tar(.gz)|db> [<dir|tar>]
# Decompressing a subset of Foldcomp database
foldcomp decompress [-t number] --id-list <idlist.txt> <db> [<dir|tar>]
# Extraction of sequence or pLDDT
foldcomp extract [--plddt|--amino-acid] <fcz> [<fasta>]
foldcomp extract [--plddt|--amino-acid] [-t number] <dir|tar(.gz)|db> [<fasta_out>]
# Check
foldcomp check <fcz>
foldcomp check [-t number] <dir|tar(.gz)|db>
# RMSD
foldcomp rmsd <pdb|cif> <pdb|cif>
# Options
-h, --help print this help message
-v, --version print version
-t, --threads threads for (de)compression of folders/tar files [default=1]
-r, --recursive recursively look for files in directory [default=0]
-f, --file input is a list of files [default=0]
-a, --alt use alternative atom order [default=false]
-b, --break interval size to save absolute atom coordinates [default=25]
-z, --tar save as tar file [default=false]
-d, --db save as database [default=false]
-y, --overwrite overwrite existing files [default=false]
-l, --id-list a file of id list to be processed (only for database input)
--skip-discontinuous skip PDB with with discontinuous residues (only batch compression)
--check check FCZ before and skip entries with error (only for batch decompression)
--plddt extract pLDDT score (only for extraction mode)
--fasta extract amino acid sequence (only for extraction mode)
--no-merge do not merge output files (only for extraction mode)
--time measure time for compression/decompression
Downloading Databases
We offer prebuilt databases for multiple large sets of predicted protein structures and a Python helper to download the database files.
You can download the AlphaFoldDB Swiss-Prot with the following command:
python -c "import foldcomp; foldcomp.setup('afdb_swissprot_v4');
Currently we offer the following databases:
-
ESMAtlas full (v0 + v2023_02):
foldcomp.setup('esmatlas') -
ESMAtlas v2023_02:
foldcomp.setup('esmatlas_v2023_02') -
ESMAtlas high-quality:
foldcomp.setup('highquality_clust30')Note: We skipped all structures with discontinous residues or other issues. Here is a list with the affected predictions; full (~21M), high-quality (~100k), v2023_02 (~10k)
-
AlphaFoldDB Uniprot:
foldcomp.setup('afdb_uniprot_v4') -
AlphaFoldDB Swiss-Prot:
foldcomp.setup('afdb_swissprot_v4') -
AlphaFoldDB Model Organisms:
foldcomp.setup('h_sapiens')a_thaliana,c_albicans,c_elegans,d_discoideum,d_melanogaster,d_rerio,e_coli,g_max,h_sapiens,m_jannaschii,m_musculus,o_sativa,r_norvegicus,s_cerevisiae,s_pombe,z_mays
-
AlphaFoldDB Cluster Representatives:
foldcomp.setup('afdb_rep_v4') -
AlphaFoldDB Cluster Representatives (Dark Clusters):
foldcomp.setup('afdb_rep_dark_v4')
If you want other prebuilt datasets, please get in touch with us through our GitHub issues.
If you have issues downloading the databases you can navigate directly to our download server and download the required files. E.g. afdb_uniprot_v4, afdb_uniprot_v4.index, afdb_uniprot_v4.dbtype, afdb_uniprot_v4.lookup, and optionally afdb_uniprot_v4.source.
Python API
You can find more in-depth examples of using Foldcomp's Python interface in the example notebook:
import foldcomp
# 01. Handling a FCZ file
# Open a fcz file
with open("test/compressed.fcz", "rb") as fcz:
fcz_binary = fcz.read()
# Decompress
(name, pdb) = foldcomp.decompress(fcz_binary) # pdb_out[0]: file name, pdb_out[1]: pdb binary string
# Save to a pdb file
with open(name, "w") as pdb_file:
pdb_file.write(pdb)
# Get data as dictionary
data_dict = foldcomp.get_data(fcz_binary) # foldcomp.get_data(pdb) also works
# Keys: phi, psi, omega, torsion_angles, residues, bond_angles, coordinates
data_dict["phi"] # phi angles (C-N-CA-C)
data_dict["psi"] # psi angles (N-CA-C-N)
data_dict["omega"] # omega angles (CA-C-N-CA)
data_dict["torsion_angles"] # torsion angles of the backbone as list (phi + psi + omega)
data_dict["bond_angles"] # bond angles of the backbone as list
data_dict["residues"] # amino acid residues as string
data_dict["coordinates"] # coordinates of the backbone as list
# 02. Iterate over a database of FCZ files
# Open a foldcomp database
ids = ["d1asha_", "d1it2a_"]
with foldcomp.open("test/example_db", ids=ids) as db:
# Iterate through database
for (name, pdb) in db:
# save entries as seperate pdb files
with open(name + ".pdb", "w") as pdb_file:
pdb_file.write(pdb)
Subsetting Databases
If you are dealing with millions of entries, we recommend using createsubdb command
of mmseqs2 to subset databases.
The following commands can be used to subset the AlphaFold Uniprot DB with given IDs.
# mmseqs createsubdb --subdb-mode 0 --id-mode 1 id_list.txt input_foldcomp_db output_foldcomp_db
mmseqs createsubdb --subdb-mode 0 --id-mode 1 id_list.txt afdb_uniprot_v4 afdb_subset
Please note that the IDs in afdb_uniprot_v4 are in the format AF-A0A5S3Y9Q7-F1-model_v4 .
Community Contributions
- PyMOL Plugin for reading Foldcomp files by @yakomaxa
Contributor

Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distributions
File details
Details for the file foldcomp-0.0.7.tar.gz.
File metadata
- Download URL: foldcomp-0.0.7.tar.gz
- Upload date:
- Size: 21.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 56e8aebab5a48d44052d533ee35dc2c8cdbe7a10472d3ade934d584e90c59b21 |
|
| MD5 | ad69fc59d51ea43215973add69e78748 |
|
| BLAKE2b-256 | 3eeaf1b28fc3e063f909fb83241aa0042940c0fa4098320b1f4855c7724f6f4e |
File details
Details for the file foldcomp-0.0.7-cp311-cp311-win_amd64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp311-cp311-win_amd64.whl
- Upload date:
- Size: 154.3 kB
- Tags: CPython 3.11, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 7997ce6219174f894666b81292011910637053d3a98a64a583ab54e78d10b1ec |
|
| MD5 | a1d78180335071b2fdb6459e3f09ed5f |
|
| BLAKE2b-256 | 32c5c64279346ebfce7664d86a3130cf22e50a43b27a37e1e44ed6f82dd4cda2 |
File details
Details for the file foldcomp-0.0.7-cp311-cp311-musllinux_1_1_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp311-cp311-musllinux_1_1_x86_64.whl
- Upload date:
- Size: 798.0 kB
- Tags: CPython 3.11, musllinux: musl 1.1+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 1946c813087de71841c92f2763b0926ce9e89f5f1882a3ed49fa6b85880921c1 |
|
| MD5 | 851e935ba702098d1e5cb7941b29458a |
|
| BLAKE2b-256 | 702f80814f26928bbffd89bc1d2ea9143b52797520fd7b831b0de12940366d82 |
File details
Details for the file foldcomp-0.0.7-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp311-cp311-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
- Upload date:
- Size: 266.6 kB
- Tags: CPython 3.11, manylinux: glibc 2.17+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 52cac8d0e9c48369ffb1f7848961afd900fe0fddca3cb07427809969cf39f575 |
|
| MD5 | e4aa3847f4cfeec42855da4faa8cb422 |
|
| BLAKE2b-256 | ee40dc68eba8e424740c1610928631653acc3f58b01f9cca26bd6e81ab7073ac |
File details
Details for the file foldcomp-0.0.7-cp311-cp311-macosx_10_9_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp311-cp311-macosx_10_9_x86_64.whl
- Upload date:
- Size: 248.0 kB
- Tags: CPython 3.11, macOS 10.9+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | b279f5ad06193b78db7fe680b05f53809e6e97f630356d3dc57ac1f5528ae8bc |
|
| MD5 | ec4b09032a829cc06cf6b711fee9e397 |
|
| BLAKE2b-256 | 6e896ccaeb1ebd392c4bd622d1c646ccc2ecf324b6e88f04dd7278d230055ab2 |
File details
Details for the file foldcomp-0.0.7-cp311-cp311-macosx_10_9_universal2.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp311-cp311-macosx_10_9_universal2.whl
- Upload date:
- Size: 473.3 kB
- Tags: CPython 3.11, macOS 10.9+ universal2 (ARM64, x86-64)
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 7aec763b930f2552871b925e043173417dfd7920accaa4e823a2eb1eb30054fe |
|
| MD5 | ddcb7225912f3ed7517a3837cb02b8b0 |
|
| BLAKE2b-256 | 2a00688fc1bfac5b1996b36a4f3e851a4198fbae8266981431e9161c16afd382 |
File details
Details for the file foldcomp-0.0.7-cp310-cp310-win_amd64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp310-cp310-win_amd64.whl
- Upload date:
- Size: 154.3 kB
- Tags: CPython 3.10, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 61eb9e35475ef454e56d74e9beddc34c67df5e0a4a0604672398e8856d203777 |
|
| MD5 | 352ac5730b62c07738dfd41b0b36fc32 |
|
| BLAKE2b-256 | 217798cff9433083256c84aa38489436fae79a7b907e49ec54d29576bd2a5735 |
File details
Details for the file foldcomp-0.0.7-cp310-cp310-musllinux_1_1_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp310-cp310-musllinux_1_1_x86_64.whl
- Upload date:
- Size: 798.0 kB
- Tags: CPython 3.10, musllinux: musl 1.1+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 218720c0ce3f0fa7088ca30bfff4ec34ef83f3afbea9b87fb9426bbee1e6c30c |
|
| MD5 | 21a14e21ea4b14c6b3022f83699de3aa |
|
| BLAKE2b-256 | b25ad95a18c8ec525ab7727d0557042c719e13e2d791bce6f3cfb5f284936ac9 |
File details
Details for the file foldcomp-0.0.7-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp310-cp310-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
- Upload date:
- Size: 266.6 kB
- Tags: CPython 3.10, manylinux: glibc 2.17+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | ff229be20e2cfffa12b5c566a4c92c9667b1546300b5669c25c21baf58cd0e4e |
|
| MD5 | 16092ecbf281190604bbfe7b956ac6c1 |
|
| BLAKE2b-256 | 33fd9f7bb10b8925750e728456af8a869ae713273e74a04afa47f749b4e5c59d |
File details
Details for the file foldcomp-0.0.7-cp310-cp310-macosx_10_9_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp310-cp310-macosx_10_9_x86_64.whl
- Upload date:
- Size: 248.0 kB
- Tags: CPython 3.10, macOS 10.9+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 8174d9d4aa43ab1e3f012b6b6eaeb1d4bab23ef7a8b9e931ae69e0c4579cb5c1 |
|
| MD5 | b34e844f0130d76e1f41c26ff62b8e2f |
|
| BLAKE2b-256 | a4dd4204d433ebf069e542c05fa870c8339011dcae2491875ebfa04b736f8bd9 |
File details
Details for the file foldcomp-0.0.7-cp310-cp310-macosx_10_9_universal2.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp310-cp310-macosx_10_9_universal2.whl
- Upload date:
- Size: 473.3 kB
- Tags: CPython 3.10, macOS 10.9+ universal2 (ARM64, x86-64)
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 780cf8abf687d25822c3d393b790540adef8f246cb226804962a79a351b95b57 |
|
| MD5 | df28d4bfb6431a9adcf401d893c9f4db |
|
| BLAKE2b-256 | e7468b306556de097b31e2cf11ead0f3f15bcf6a84fda3e8004e3d8a08df7a0c |
File details
Details for the file foldcomp-0.0.7-cp39-cp39-win_amd64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp39-cp39-win_amd64.whl
- Upload date:
- Size: 154.3 kB
- Tags: CPython 3.9, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 107a5e7151d90cda00fbd20eff51cd8cf88e6f87d48704db78dd457770a2ccc3 |
|
| MD5 | e2fffc99925f61774265afb93e564ddc |
|
| BLAKE2b-256 | c5b247c142d33bb802dd1505c24ce43b08ebf396e53891f5400f5e0ad062dd66 |
File details
Details for the file foldcomp-0.0.7-cp39-cp39-musllinux_1_1_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp39-cp39-musllinux_1_1_x86_64.whl
- Upload date:
- Size: 798.0 kB
- Tags: CPython 3.9, musllinux: musl 1.1+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 009956e7678045a7f49e2fa7c5ea30311090107ae0f562dd8bd4037fd53fdb32 |
|
| MD5 | 5fc7786a896f7f3ae2066c1cf4c3e1de |
|
| BLAKE2b-256 | 679d4d6d3f03525f173380abb347e12b7c62b7006583c614079742e4f8dd90c9 |
File details
Details for the file foldcomp-0.0.7-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp39-cp39-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
- Upload date:
- Size: 266.6 kB
- Tags: CPython 3.9, manylinux: glibc 2.17+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | f41aff4c18399811a2571f3d999a9abb33c681fcb0cbb96d87f3c2f7616235bc |
|
| MD5 | 071341c4502efe8ea737da3c62aca863 |
|
| BLAKE2b-256 | ece69f70dfae43c2afc1b197dcae2d80006550f29a44563a8776c5ae704321f1 |
File details
Details for the file foldcomp-0.0.7-cp39-cp39-macosx_10_9_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp39-cp39-macosx_10_9_x86_64.whl
- Upload date:
- Size: 248.0 kB
- Tags: CPython 3.9, macOS 10.9+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 83401f32dc996ec26314691ab5e65ed362d4adcd2c2935cbef2996cdf47e8038 |
|
| MD5 | 144c95c616c67bca4869bdf4097b46c7 |
|
| BLAKE2b-256 | f55bf783e68dce29530a192e23b8111f373a3128fe9532b63df9a4d2653036a0 |
File details
Details for the file foldcomp-0.0.7-cp39-cp39-macosx_10_9_universal2.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp39-cp39-macosx_10_9_universal2.whl
- Upload date:
- Size: 473.3 kB
- Tags: CPython 3.9, macOS 10.9+ universal2 (ARM64, x86-64)
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 24eb2ba08e6fe908e4b239a6ba590ec54d1b851620e419744df6e697bdf36a66 |
|
| MD5 | e7e97b2e5fd46bd4fe30baf2e02b0e97 |
|
| BLAKE2b-256 | 04ca0aa8d6f325610ec0054b9e89d8deab7560f33a3e1345a208deb48b4685d9 |
File details
Details for the file foldcomp-0.0.7-cp38-cp38-win_amd64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp38-cp38-win_amd64.whl
- Upload date:
- Size: 154.3 kB
- Tags: CPython 3.8, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 98a4c7f199dcc4afda8dff00c7861d8d8774b5133ec474a678d999deb7a7e1eb |
|
| MD5 | 88b64fbcc9007b45818283257db1f6c5 |
|
| BLAKE2b-256 | 4092416c2560bd52caf3ae9eb1746990e422a1805428e351062eecf11c3ab0ce |
File details
Details for the file foldcomp-0.0.7-cp38-cp38-musllinux_1_1_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp38-cp38-musllinux_1_1_x86_64.whl
- Upload date:
- Size: 798.0 kB
- Tags: CPython 3.8, musllinux: musl 1.1+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 0c797e533570f1e03ce926200de50652880c4d1b266c12d8b297a2e7f16827c8 |
|
| MD5 | 36f1b228d21bc704b5efee8f6cbeea08 |
|
| BLAKE2b-256 | 5a65f373340cb0565e660501b59bc8007e3c14b86966d424d4e07ef9b5dcf23d |
File details
Details for the file foldcomp-0.0.7-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp38-cp38-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
- Upload date:
- Size: 266.6 kB
- Tags: CPython 3.8, manylinux: glibc 2.17+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 8e3efee2e1ba4661ed376b6436ba88bd0c90037698de5d4085fe6ab64dfb552f |
|
| MD5 | 366d02fa007e92dc2dbc5c090bdc172c |
|
| BLAKE2b-256 | 00535e0f03e50e2dc0c07f99daf21d9edd68926a16357d4e9be0b3d1ceedc203 |
File details
Details for the file foldcomp-0.0.7-cp38-cp38-macosx_10_9_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp38-cp38-macosx_10_9_x86_64.whl
- Upload date:
- Size: 248.0 kB
- Tags: CPython 3.8, macOS 10.9+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | facaf4d301ab49dc97658dd80939995f8722f9c21a66ddc39024fb692ea8d5e6 |
|
| MD5 | 0d6384c3e585d8c52c92c851dfa9cfa9 |
|
| BLAKE2b-256 | 3d1dc1b0a634f6c1c9469acad47983cc2ad282dbd8d852b979e12a345e57cf70 |
File details
Details for the file foldcomp-0.0.7-cp38-cp38-macosx_10_9_universal2.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp38-cp38-macosx_10_9_universal2.whl
- Upload date:
- Size: 473.3 kB
- Tags: CPython 3.8, macOS 10.9+ universal2 (ARM64, x86-64)
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | bfbaf1f60fd523067b51bdada473060d9146848c0efb78d39109ac9fed572707 |
|
| MD5 | f2284b733af0fe6d4d379080751d4439 |
|
| BLAKE2b-256 | d7be5c2a9fb830afa3be5910b0933c80c6e1f97f10018207da027d5673f92c70 |
File details
Details for the file foldcomp-0.0.7-cp37-cp37m-win_amd64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp37-cp37m-win_amd64.whl
- Upload date:
- Size: 154.2 kB
- Tags: CPython 3.7m, Windows x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 081fadbb968eb1b11603df48435dbf124832b407c5e556756ff7073ad78fb8a4 |
|
| MD5 | 3a6c460267e8c7a5cf4a4d73c5ba4a76 |
|
| BLAKE2b-256 | c78d972bbdb49dc38ece096f8f0c1b07a8aa27a92b7ffc011c4fe1a0acafa1cb |
File details
Details for the file foldcomp-0.0.7-cp37-cp37m-musllinux_1_1_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp37-cp37m-musllinux_1_1_x86_64.whl
- Upload date:
- Size: 797.7 kB
- Tags: CPython 3.7m, musllinux: musl 1.1+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | be7daf7e0c203f81953c209372655427e1800f140746ea569d0b861ae8883f9d |
|
| MD5 | 05a6d560de0b75147db7fcfa372a363a |
|
| BLAKE2b-256 | 934188753c31bef3d42666ed4efb45fe0f1308a345358d5966165f1c8347ef89 |
File details
Details for the file foldcomp-0.0.7-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp37-cp37m-manylinux_2_17_x86_64.manylinux2014_x86_64.whl
- Upload date:
- Size: 266.5 kB
- Tags: CPython 3.7m, manylinux: glibc 2.17+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 6e57601144439646f9a28fc1a981b8dfd4498d7e7630e24dd9eee0cd8295a3f9 |
|
| MD5 | 6ad9fec25ba5d39708cdd8448e11cefb |
|
| BLAKE2b-256 | 005dcd94c0bb832720fa1357cd155e267d60c4719e3c6c0f2ae2b3a98b1ddc58 |
File details
Details for the file foldcomp-0.0.7-cp37-cp37m-macosx_10_9_x86_64.whl.
File metadata
- Download URL: foldcomp-0.0.7-cp37-cp37m-macosx_10_9_x86_64.whl
- Upload date:
- Size: 248.2 kB
- Tags: CPython 3.7m, macOS 10.9+ x86-64
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.18
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 629270d531c2187307ba6159af6085808ea646565b67d03718bda5109953b9b1 |
|
| MD5 | 88d66a3f629b5ef880941f0eb628be8b |
|
| BLAKE2b-256 | 285380f9a776b06439e28296a2111e43362d4a4511d76811bde192da0b2ab0f2 |












