Image Based Ecological Information System
Project description

This project is a component of the WildMe / WildBook project: See https://github.com/WildbookOrg/
IBEIS - Image Analysis
I.B.E.I.S. = Image Based Ecological Information System

Program Description
IBEIS program for the storage and management of images and derived data for use in computer vision algorithms. It aims to compute who an animal is, what species an animal is, and where an animal is with the ultimate goal being to ask important why biological questions. This This repo Image Analysis image analysis module of IBEIS. It is both a python module and standalone program.
Currently the system is build around and SQLite database, a PyQt4 GUI, and matplotlib visualizations. Algorithms employed are: random forest species detection and localization, hessian-affine keypoint detection, SIFT keypoint description, LNBNN identification using approximate nearest neighbors. Algorithms in development are SMK (selective match kernel) for identification and deep neural networks for detection and localization.
The core of IBEIS is the IBEISController class. It provides an API into IBEIS data management and algorithms. The IBEIS API Documentation can be found here: http://erotemic.github.io/ibeis
The IBEIS GUI (graphical user interface) is built on top of the API. We are also experimenting with a new web frontend that bypasses the older GUI code.
## Self Installing Executables:
Unfortunately we have not released self-installing-executables for IBEIS yet. We plan to release these “soon”.
However there are old HotSpotter (the software which IBEIS is based on) binaries available. These can be downloaded from: http://cs.rpi.edu/hotspotter/
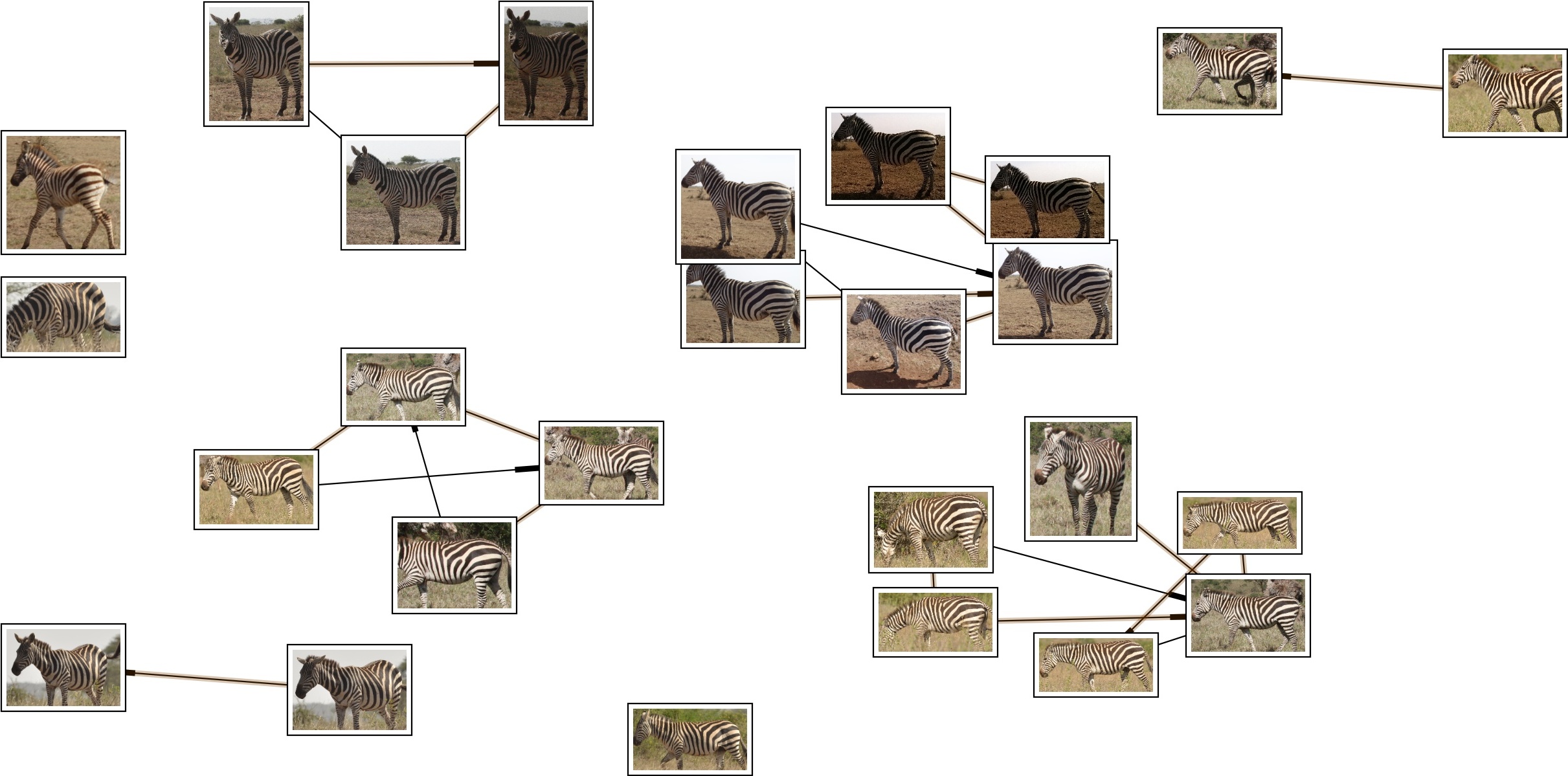
Visual Demo
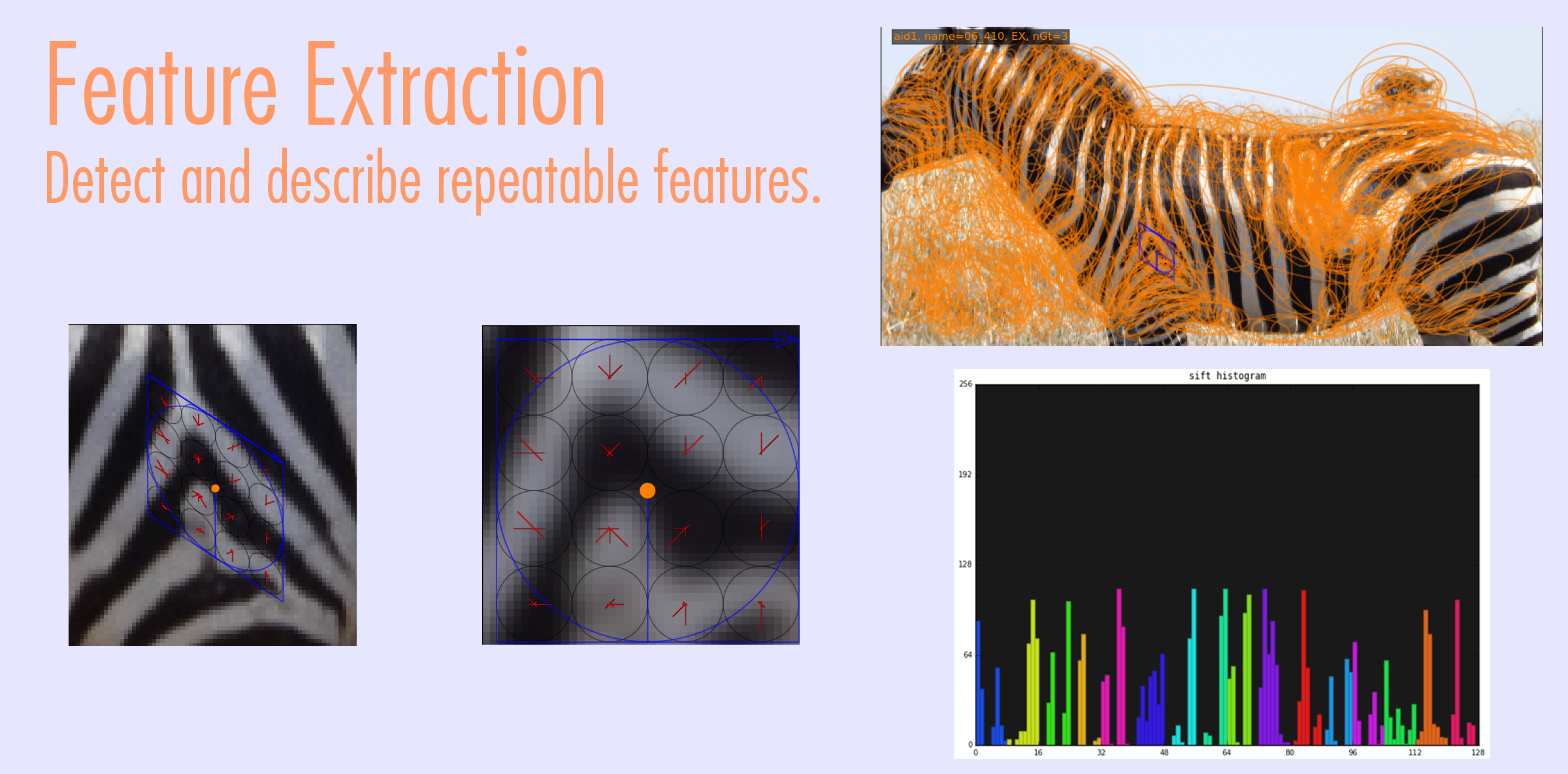
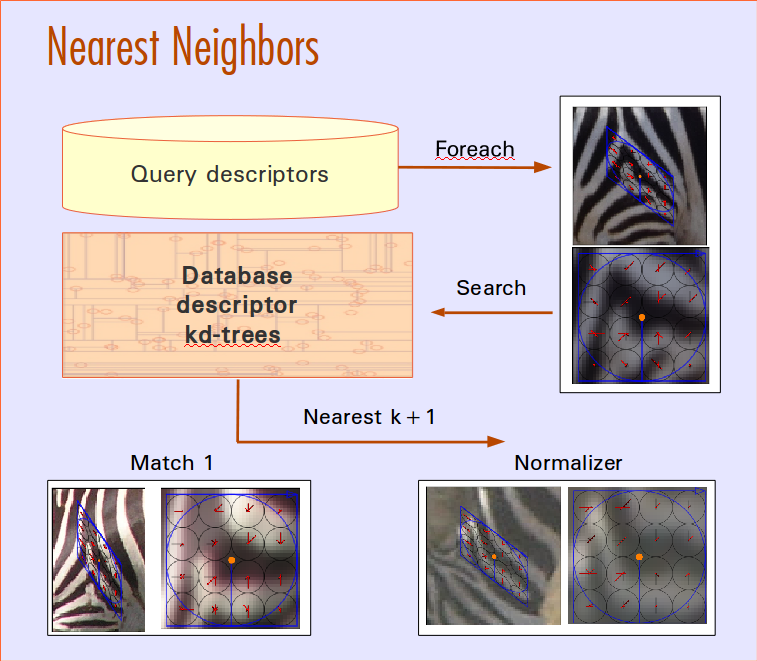
Match Scoring
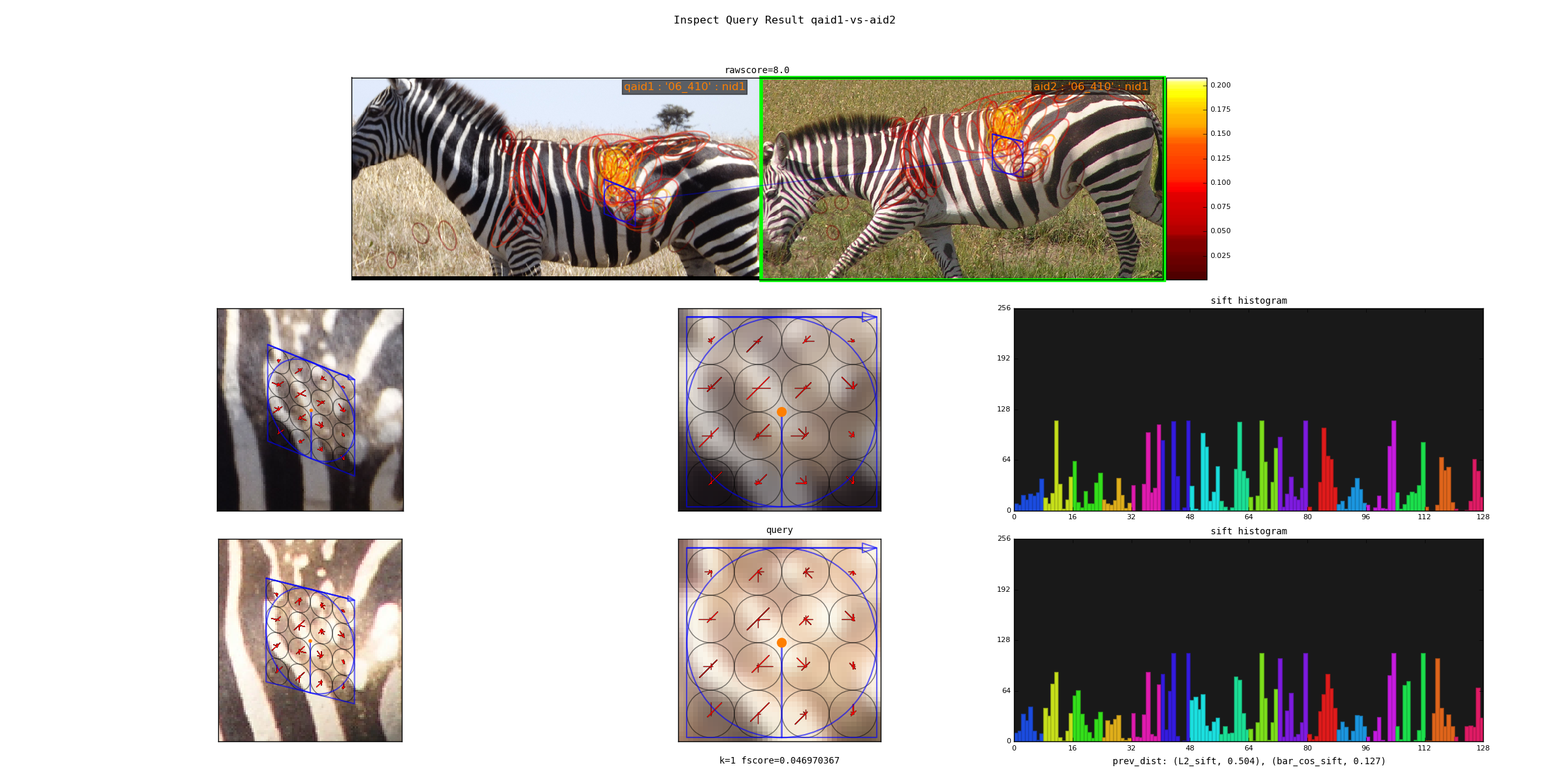
Spatial Verification

python -m vtool.spatial_verification --test-spatially_verify_kpts --showName Scoring

python -m ibeis.algo.hots.chip_match show_single_namematch --qaid 1 --showIdentification Ranking

python -m ibeis.algo.hots.chip_match show_ranked_matches --show --qaid 86Inference
# broken
# python -m ibeis.algo.preproc.preproc_encounter compute_encounter_groups --showInternal Modules
In the interest of modular code we are actively developing several different modules.
Erotemic’s IBEIS Image Analysis module dependencies
bluemellophone’s IBEIS Image Analysis modules
The IBEIS module itself:
IBEIS Development Environment Setup
NOTE: this section is outdated.
# The following install script install ibeis and all dependencies.
# If it doesnt you can look at the older instructions which follow
# and try to figure it out. After running this you should have a code
# directory with all of the above repos.
# Navigate to your code directory
export CODE_DIR=~/code
mkdir $CODE_DIR
cd $CODE_DIR
# Clone IBEIS
git clone https://github.com/Erotemic/ibeis.git
cd ibeis
# Install the requirements for super_setup
pip install -r requirements/super_setup.txt
# Install the development requirements (note-these are now all on pypi, so
# this is not strictly necessary)
python super_setup.py ensure
# NOTE: you can use super_setup to do several things
python super_setup.py --help
python super_setup.py versions
python super_setup.py status
python super_setup.py check
python super_setup.py pull
# Run the run_developer_setup.sh file in each development repo
python super_setup.py develop
# Or you can also just do to use pypi versions of dev repos:
python setup.py develop
# Optional: set a workdir and download a test dataset
.python -m ibeis.dev
.python -m ibeis.dev -t mtest
python -m ibeis.dev -t nauts
./reset_dbs.py
python -m ibeis --set-workdir ~/data/work --preload-exit
python -m ibeis -e ensure_mtest
# make sure everyhing is set up correctly
python -m ibeis --db PZ_MTESTRunning Tests
The new way of running tests is with xdoctest, or using the “run_doctests.sh” script.
Example usage
(Note: This list is far from complete)
#--------------------
# Main Commands
#--------------------
python -m ibeis.main <optional-arguments> [--help]
python -m ibeis.dev <optional-arguments> [--help]
# main is the standard entry point to the program
# dev is a more advanced developer entry point
# ** NEW 7-23-2015 **: the following commands are now equivalent and do not
# have to be specified from the ibeis source dir if ibeis is installed
python -m ibeis <optional-arguments> [--help]
python -m ibeis.dev <optional-arguments> [--help]
# Useful flags.
# Read code comments in dev.py for more info.
# Careful some commands don't work. Most do.
# --cmd # shows ipython prompt with useful variables populated
# -w, --wait # waits (useful for showing plots)
# --gui # starts the gui as well (dev.py does not show gui by default, main does)
# --web # runs the program as a web server
# --quiet # turns off most prints
# --verbose # turns on verbosity
# --very-verbose # turns on extra verbosity
# --debug2 # runs extra checks
# --debug-print # shows where print statments occur
# -t [test]
#--------------------
# PSA: Workdirs:
#--------------------
# IBEIS uses the idea of a work directory for databases.
# Use --set-workdir <path> to set your own, or a gui will popup and ask you about it
./main.py --set-workdir /raid/work --preload-exit
./main.py --set-logdir /raid/logs/ibeis --preload-exit
python -m ibeis.dev --set-workdir ~/data/work --preload-exit
# use --db to specify a database in your WorkDir
# --setdb makes that directory your default directory
python -m ibeis.dev --db <dbname> --setdb
# Or just use the absolute path
python -m ibeis.dev --dbdir <full-dbpath>
#--------------------
# Examples:
# Here are are some example commands
#--------------------
# Run the queries for each roi with groundtruth in the PZ_MTEST database
# using the best known configuration of parameters
python -m ibeis.dev --db PZ_MTEST --allgt -t best
python -m ibeis.dev --db PZ_MTEST --allgt -t score
# View work dir
python -m ibeis.dev --vwd --prequit
# List known databases
python -m ibeis.dev -t list_dbs
# Dump/Print contents of params.args as a dict
python -m ibeis.dev --prequit --dump-argv
# Dump Current SQL Schema to stdout
python -m ibeis.dev --dump-schema --postquit
#------------------
# Convert a hotspotter database to IBEIS
#------------------
# NEW: You can simply open a hotspotter database and it will be converted to IBEIS
python -m ibeis convert_hsdb_to_ibeis --dbdir <path_to_hsdb>
# This script will exlicitly conver the hsdb
python -m ibeis convert_hsdb_to_ibeis --hsdir <path_to_hsdb> --dbdir <path_to_newdb>
#---------
# Ingest examples
#---------
# Ingest raw images
python -m ibeis.dbio.ingest_database --db JAG_Kieryn
#---------
# Run Tests
#---------
./run_tests.py
#----------------
# Test Commands
#----------------
# Set a default DB First
python -m ibeis.dev --setdb --dbdir /path/to/your/DBDIR
python -m ibeis.dev --setdb --db YOURDB
python -m ibeis.dev --setdb --db PZ_MTEST
python -m ibeis.dev --setdb --db PZ_FlankHack
# List all available tests
python -m ibeis.dev -t help
# Minimal Database Statistics
python -m ibeis.dev --allgt -t info
# Richer Database statistics
python -m ibeis.dev --allgt -t dbinfo
# Print algorithm configurations
python -m ibeis.dev -t printcfg
# Print database tables
python -m ibeis.dev -t tables
# Print only the image table
python -m ibeis.dev -t imgtbl
# View data directory in explorer/finder/nautilus
python -m ibeis.dev -t vdd
# List all IBEIS databases
python -m ibeis list_dbs
# Delete cache
python -m ibeis delete_cache --db testdb1
# Show a single annotations
python -m ibeis.viz.viz_chip show_chip --db PZ_MTEST --aid 1 --show
# Show annotations 1, 3, 5, and 11
python -m ibeis.viz.viz_chip show_many_chips --db PZ_MTEST --aids=1,3,5,11 --show
# Database Stats for all our important datasets:
python -m ibeis.dev --allgt -t dbinfo --db PZ_MTEST | grep -F "[dbinfo]"
# Some mass editing of metadata
python -m ibeis.dev --db PZ_FlankHack --edit-notes
python -m ibeis.dev --db GZ_Siva --edit-notes
python -m ibeis.dev --db GIR_Tanya --edit-notes
python -m ibeis.dev --allgt -t dbinfo --db GZ_ALL --set-all-species zebra_grevys
# Current Experiments:
# Main experiments
python -m ibeis --tf draw_annot_scoresep --db PZ_MTEST -a default -t best --show
python -m ibeis.dev -e draw_rank_cdf --db PZ_MTEST --show -a timectrl
# Show disagreement cases
ibeis --tf draw_match_cases --db PZ_MTEST -a default:size=20 \
-t default:K=[1,4] \
--filt :disagree=True,index=0:4 --show
# SMK TESTS
python -m ibeis.dev -t smk2 --allgt --db PZ_MTEST --nocache-big --nocache-query --qindex 0:20
python -m ibeis.dev -t smk2 --allgt --db PZ_MTEST --qindex 20:30 --va
# Feature Tuning
python -m ibeis.dev -t test_feats -w --show --db PZ_MTEST --allgt --qindex 1:2
python -m ibeis.dev -t featparams -w --show --db PZ_MTEST --allgt
python -m ibeis.dev -t featparams_big -w --show --db PZ_MTEST --allgt
# NEW DATABASE TEST
python -m ibeis.dev -t best --db seals2 --allgt
# Testing Distinctivness Parameters
python -m ibeis.algo.hots.distinctiveness_normalizer --test-get_distinctiveness --show --db GZ_ALL --aid 2
python -m ibeis.algo.hots.distinctiveness_normalizer --test-get_distinctiveness --show --db PZ_MTEST --aid 10
python -m ibeis.algo.hots.distinctiveness_normalizer --test-test_single_annot_distinctiveness_params --show --db GZ_ALL --aid 2
# 2D Gaussian Curves
python -m vtool_ibeis.patch --test-test_show_gaussian_patches2 --show
# Test Keypoint Coverage
python -m vtool_ibeis.coverage_kpts --test-gridsearch_kpts_coverage_mask --show
python -m vtool_ibeis.coverage_kpts --test-make_kpts_coverage_mask --show
# Test Grid Coverage
python -m vtool_ibeis.coverage_grid --test-gridsearch_coverage_grid_mask --show
python -m vtool_ibeis.coverage_grid --test-sparse_grid_coverage --show
python -m vtool_ibeis.coverage_grid --test-gridsearch_coverage_grid --show
# Test Spatially Constrained Scoring
python -m ibeis.algo.hots.vsone_pipeline --test-compute_query_constrained_matches --show
python -m ibeis.algo.hots.vsone_pipeline --test-gridsearch_constrained_matches --show
# Test VsMany ReRanking
python -m ibeis.algo.hots.vsone_pipeline --test-vsone_reranking --show
python -m ibeis.algo.hots.vsone_pipeline --test-vsone_reranking --show --homog
# Problem cases with the back spot
python -m ibeis.algo.hots.vsone_pipeline --test-vsone_reranking --show --homog --db GZ_ALL --qaid 425
python -m ibeis.algo.hots.vsone_pipeline --test-vsone_reranking --show --homog --db GZ_ALL --qaid 662
python -m ibeis.dev -t custom:score_method=csum,prescore_method=csum --db GZ_ALL --show --va -w --qaid 425 --noqcache
# Shows vsone results with some of the competing cases
python -m ibeis.algo.hots.vsone_pipeline --test-vsone_reranking --show --homog --db GZ_ALL --qaid 662 --daid_list=425,342,678,233
# More rerank vsone tests
python -c "import utool as ut; ut.write_modscript_alias('Tbig.sh', 'dev.py', '--allgt --db PZ_Master0')"
sh Tbig.sh -t custom:rrvsone_on=True custom
sh Tbig.sh -t custom:rrvsone_on=True custom --noqcache
#----
# Turning back on name scoring and feature scoring and restricting to rerank a subset
# This gives results that are closer to what we should actually expect
python -m ibeis.dev --allgt -t custom \
custom:rrvsone_on=True,prior_coeff=1.0,unconstrained_coeff=0.0,fs_lnbnn_min=0,fs_lnbnn_max=1 \
custom:rrvsone_on=True,prior_coeff=0.5,unconstrained_coeff=0.5,fs_lnbnn_min=0,fs_lnbnn_max=1 \
custom:rrvsone_on=True,prior_coeff=0.1,unconstrained_coeff=0.9,fs_lnbnn_min=0,fs_lnbnn_max=1 \
--print-bestcfg
#----
#----
# VsOneRerank Tuning: Tune linar combination
python -m ibeis.dev --allgt -t \
custom:fg_weight=0.0 \
\
custom:rrvsone_on=True,prior_coeff=1.0,unconstrained_coeff=0.0,fs_lnbnn_min=0.0,fs_lnbnn_max=1.0,nAnnotPerNameVsOne=200,nNameShortlistVsone=200 \
\
custom:rrvsone_on=True,prior_coeff=.5,unconstrained_coeff=0.5,fs_lnbnn_min=0.0,fs_lnbnn_max=1.0,nAnnotPerNameVsOne=200,nNameShortlistVsone=200 \
\
--db PZ_MTEST
#--print-confusion-stats --print-gtscore
#----
# Testing no affine invaraiance and rotation invariance
python -m ibeis.dev -t custom:AI=True,RI=True custom:AI=False,RI=True custom:AI=True,RI=False custom:AI=False,RI=False --db PZ_MTEST --showCaveats / Things we are not currently doing
We do not add or remove points from kdtrees. They are always rebuilt
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
File details
Details for the file ibeis-2.2.2.tar.gz.
File metadata
- Download URL: ibeis-2.2.2.tar.gz
- Upload date:
- Size: 1.3 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.13.0 pkginfo/1.5.0.1 requests/2.22.0 setuptools/41.0.1 requests-toolbelt/0.9.1 tqdm/4.32.2 CPython/3.7.3
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 1f8320425e5483540434efc825f9ac69af6de6788014ed7b8215cb9c818f576a |
|
| MD5 | 71e706bec3cd579e1611efca6b339e68 |
|
| BLAKE2b-256 | fbb4c1a5195de89d4430186441fe808abd7f1c269792806e2f76af8f85df1e21 |


















