MatGL (Materials Graph Library) is a framework for graph deep learning for materials science.
Project description
MatGL (Materials Graph Library)
Table of Contents
- Introduction
- Status
- Architectures
- Installation
- Usage
- API Docs
- Developer's Guide
- References
- FAQs
- Acknowledgements
Introduction
MatGL (Materials Graph Library) is a graph deep learning library for materials science. Mathematical graphs are a natural representation for a collection of atoms (e.g., molecules or crystals). Graph deep learning models have been shown to consistently deliver exceptional performance as surrogate models for the prediction of materials properties.
In this repository, we have reimplemented the original Tensorflow MatErials 3-body Graph Network (m3gnet) and its predecessor, MEGNet, using the Deep Graph Library (DGL) and PyTorch. The goal is to improve the usability, extensibility and scalability of these models. Here are some key improvements over the TF implementations:
- A more intuitive API and class structure based on DGL.
- Multi-GPU support via PyTorch Lightning. A training utility module has been developed.
This effort is a collaboration between the Materials Virtual Lab and Intel Labs (Santiago Miret, Marcel Nassar, Carmelo Gonzales). Please refer to the official documentation for more details.
Status
Major milestones are summarized below. Please refer to change log for details.
- v0.5.1 (Jun 9 2023): Model versioning implemented.
- v0.5.0 (Jun 8 2023): Simplified saving and loading of models. Now models can be loaded with one line of code!
- v0.4.0 (Jun 7 2023): Near feature parity with original TF implementations. Re-trained M3Gnet universal potential now available.
- v0.1.0 (Feb 16 2023): Initial implementations of M3GNet and MEGNet architectures have been completed. Expect bugs!
Architectures
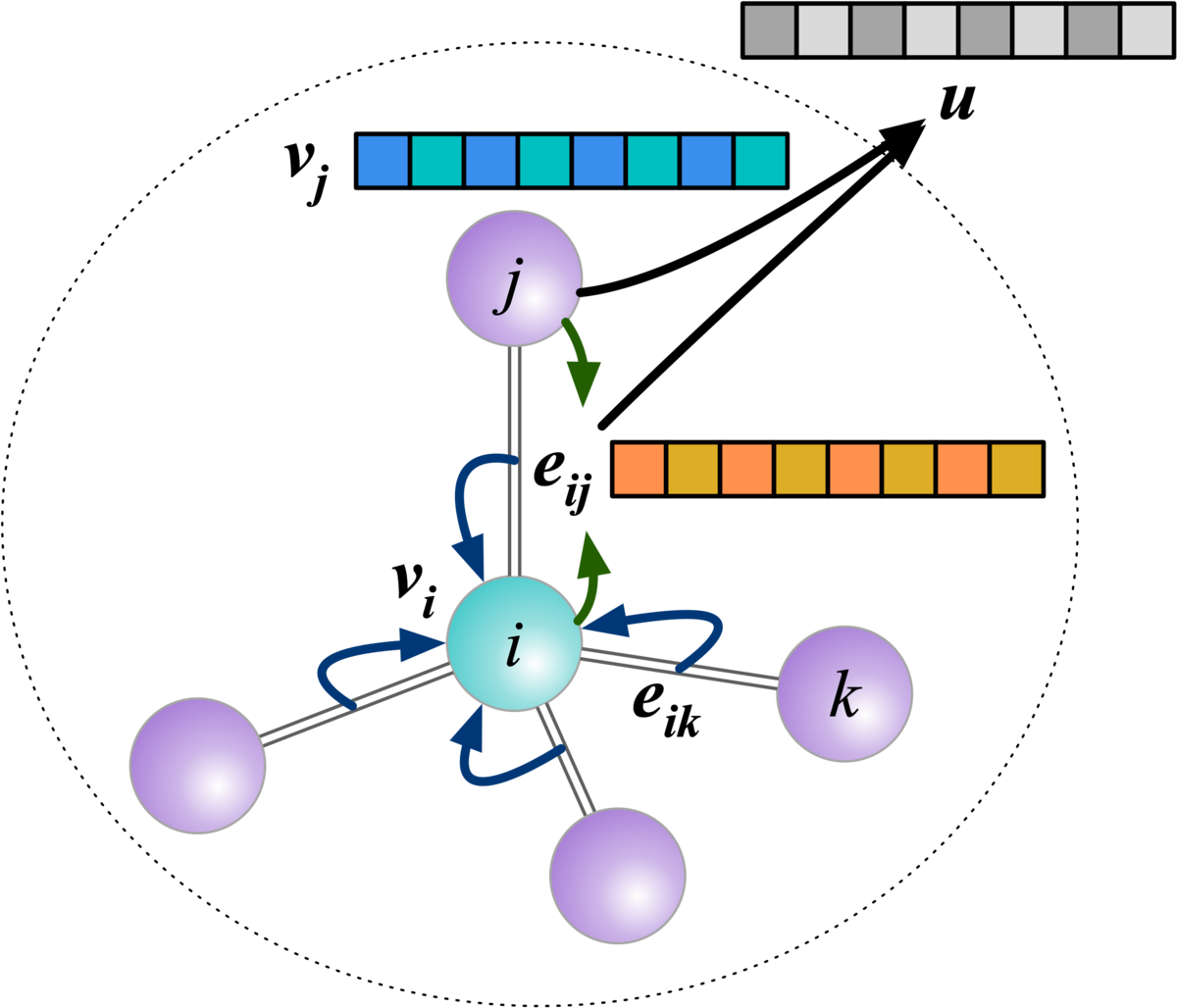
MEGNet
The MatErials Graph Network (MEGNet) is an implementation of DeepMind's graph networks for machine learning in materials science. We have demonstrated its success in achieving low prediction errors in a broad array of properties in both molecules and crystals. New releases have included our recent work on multi-fidelity materials property modeling. Figure 1 shows the sequential update steps of the graph network, whereby bonds, atoms, and global state attributes are updated using information from each other, generating an output graph.
M3GNet
M3GNet is a new materials graph neural network architecture that incorporates 3-body interactions in MEGNet. An additional difference is the addition of the coordinates for atoms and the 3×3 lattice matrix in crystals, which are necessary for obtaining tensorial quantities such as forces and stresses via auto-differentiation. As a framework, M3GNet has diverse applications, including:
- Interatomic potential development. With the same training data, M3GNet performs similarly to state-of-the-art machine learning interatomic potentials (MLIPs). However, a key feature of a graph representation is its flexibility to scale to diverse chemical spaces. One of the key accomplishments of M3GNet is the development of a universal IP that can work across the entire periodic table of the elements by training on relaxations performed in the Materials Project.
- Surrogate models for property predictions. Like the previous MEGNet architecture, M3GNet can be used to develop surrogate models for property predictions, achieving in many cases accuracies that are better or similar to other state-of-the-art ML models.
For detailed performance benchmarks, please refer to the publications in the References section.
Installation
Matgl can be installed via pip for the latest stable version:
pip install matgl
For the latest dev version, please clone this repo and install using:
python setup.py -e .
Usage
Pre-trained M3GNet universal potential and MEGNet models for the Materials Project formation energy and multi-fidelity band gap are now available. Users who just want to use the models out of the box should use the newly implemented convenience method:
import matgl
model = matgl.load_model("<model_name>")
The following is an example of a prediction of the formation energy for CsCl.
from pymatgen.core import Lattice, Structure
import matgl
model = matgl.load_model("MEGNet-MP-2018.6.1-Eform")
# This is the structure obtained from the Materials Project.
struct = Structure.from_spacegroup("Pm-3m", Lattice.cubic(4.1437), ["Cs", "Cl"], [[0, 0, 0], [0.5, 0.5, 0.5]])
eform = model.predict_structure(struct)
print(f"The predicted formation energy for CsCl is {float(eform.numpy()):.3f} eV/atom.")
Jupyter notebooks
We have written several Jupyter notebooks on the use of MatGL. These notebooks can be run on Google Colab. This will be the primary form of usage documentation.
API Docs
The Sphinx-generated API docs are available here.
Developer's Guide
A basic developer's guide has been written to outline the key design elements of matgl.
References
Please cite the following works:
MEGNet
Chen, C.; Ye, W.; Zuo, Y.; Zheng, C.; Ong, S. P. Graph Networks as a Universal Machine Learning Framework for Molecules and Crystals. Chem. Mater. 2019, 31 (9), 3564–3572. https://doi.org/10.1021/acs.chemmater.9b01294.
Multi-fidelity MEGNet
Chen, C.; Zuo, Y.; Ye, W.; Li, X.; Ong, S. P. Learning Properties of Ordered and Disordered Materials from Multi-Fidelity Data. Nature Computational Science 2021, 1, 46–53. https://doi.org/10.1038/s43588-020-00002-x.
M3GNet
Chen, C., Ong, S.P. A universal graph deep learning interatomic potential for the periodic table. Nat Comput Sci, 2, 718–728 (2022). https://doi.org/10.1038/s43588-022-00349-3.
FAQs
-
The
M3GNet-MP-2021.2.8-PESdiffers from the original TF implementation!Answer:
M3GNet-MP-2021.2.8-PESis a refitted model with some data improvements and minor architectural changes. It is not expected to reproduce the original TF implementation exactly. We have conducted reasonable benchmarks to ensure that the new implementation reproduces the broad error characteristics of the original TF implementation (see examples). It is meant to serve as a baseline for future model improvements. -
I am getting errors with
matgl.load_model()!Answer: The most likely reason is that you have an old version of the model cached. Refactoring models is common to ensure the best implementation. This can usually be solved by clearing your cache using:
python -c "import matgl; matgl.clear_cache()"
Acknowledgments
This work was primarily supported by the Materials Project, funded by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Materials Sciences and Engineering Division under contract no. DE-AC02-05-CH11231: Materials Project program KC23MP. This work used the Expanse supercomputing cluster at the Extreme Science and Engineering Discovery Environment (XSEDE), which is supported by National Science Foundation grant number ACI-1548562.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file matgl-0.5.3.tar.gz.
File metadata
- Download URL: matgl-0.5.3.tar.gz
- Upload date:
- Size: 173.2 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.16
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 1deaa8f611dde074ca819b6cbeebde582232a5fff725dd94c8419086b068b1db |
|
| MD5 | eb51d91c8d3bc20398a6195abaa051a5 |
|
| BLAKE2b-256 | 03933660076d104c784bf05ca8620ab7463030ccde13706de0c91af6a94c6a09 |
File details
Details for the file matgl-0.5.3-py3-none-any.whl.
File metadata
- Download URL: matgl-0.5.3-py3-none-any.whl
- Upload date:
- Size: 176.0 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.16
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 0a748c7a213f74eb5bd0cc3619fe89ef3b554c26213ade1004c93c7c5b5055c1 |
|
| MD5 | fab90b43893875141a5118043e96c171 |
|
| BLAKE2b-256 | 4e6267ff7d10fd4432c6a22c951a6130d59f480f3f3b1880dc136bcba42b2a2e |















