A napari plugin for interacting with electron cryotomograms
Project description
napari-tomoslice
A napari plugin for visualising and interacting with electron cryotomograms.
Installation
You can install napari-tomoslice via pip:
pip install napari-tomoslice
Usage
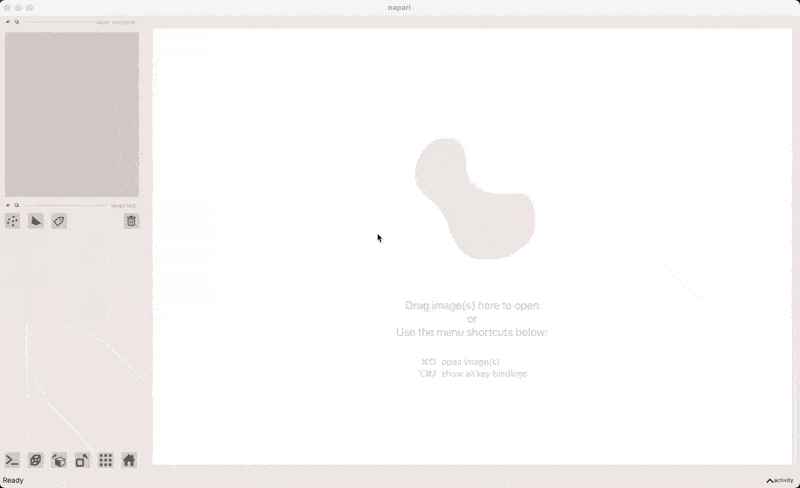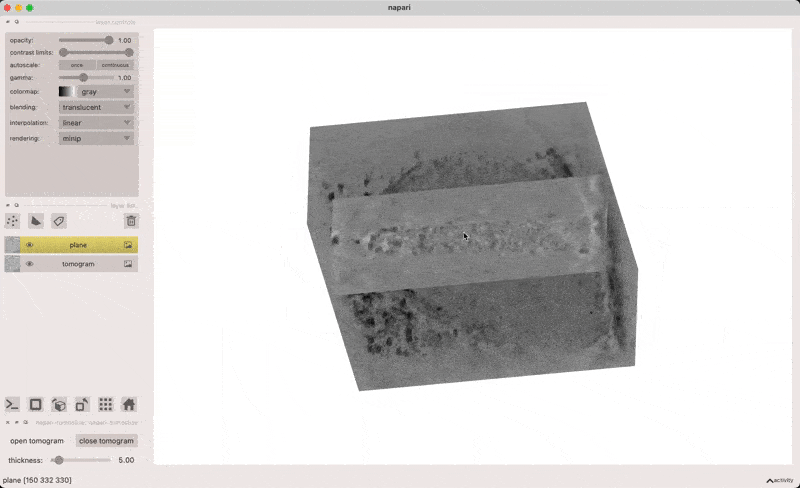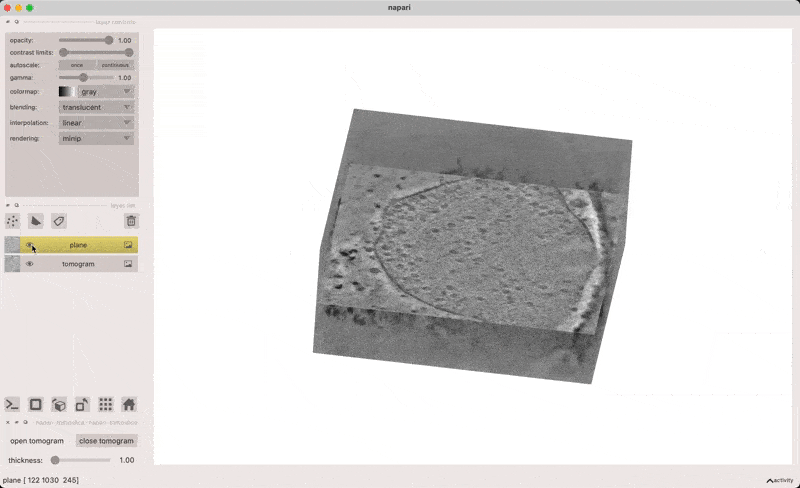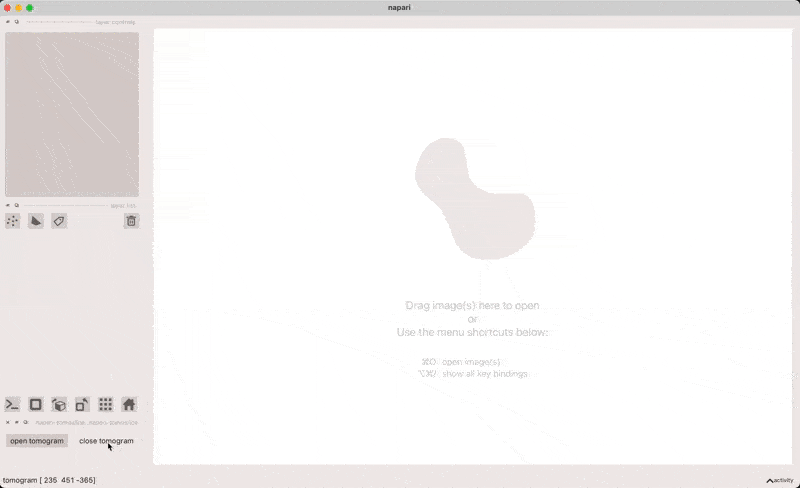
This plugin provides a user interface for opening electron cryotomograms in napari as both volumes and slices through volumes.
The plugin can be opened from the plugins menu in napari, or with
napari-tomoslice at the command line.
Usage: napari-tomoslice [TOMOGRAM_FILE]
An interactive tomogram slice viewer in napari.
Controls:
x/y/z - align normal vector along x/y/z axis
click and drag - shift plane along its normal vector
alt-click - add point on plane (if points layer is active)
o - align plane normal to view direction
[] - decrease/increase plane thickness
Arguments:
[TOMOGRAM_FILE]
Options:
--help Show this message and exit.
Contributing
Contributions are very welcome.
License
Distributed under the terms of the BSD-3 license, "napari-tomoslice" is free and open source software
Issues
If you encounter any problems, please file an issue along with a detailed description.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distributions
Built Distribution
File details
Details for the file napari_tomoslice-0.0.5-py3-none-any.whl.
File metadata
- Download URL: napari_tomoslice-0.0.5-py3-none-any.whl
- Upload date:
- Size: 15.8 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.8.1 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.9.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | f1faeb1bc5f24d72580461731b3e630c3b6bb3ef4d6c7626825ace22afb11ed6 |
|
| MD5 | 5f6645fce379b11b1a82e6cb7dc4ffee |
|
| BLAKE2b-256 | 85af14b8303e07edc00a7c9601468d9680d71c8845e8101bb652963401eeb96f |