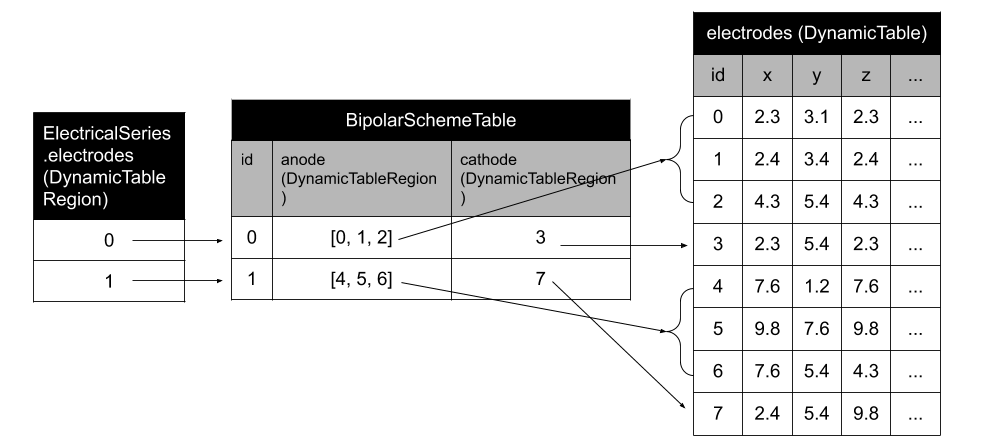
An NWB extension for storing bipolar scheme
Project description
ndx-bipolar-scheme Extension for NWB
Structure for storing the bipolar schema of a recording in an NWB file.
python installation
$ pip install ndx-bipolar-scheme
python usage
from pynwb import NWBHDF5IO, NWBFile
from pynwb.file import DynamicTableRegion
from datetime import datetime
from ndx_bipolar_scheme import BipolarSchemeTable, EcephysExt
from pynwb.ecephys import ElectricalSeries
import numpy as np
nwbfile = NWBFile('description', 'id', datetime.now().astimezone())
device = nwbfile.create_device('device_name')
electrode_group = nwbfile.create_electrode_group('electrode_group',
'desc', 'loc', device=device)
for i in np.arange(20.):
nwbfile.add_electrode(i, i, i, np.nan, 'loc', 'filt', electrode_group)
bipolar_scheme_table = BipolarSchemeTable(name='bipolar_scheme_table',
description='desc')
bipolar_scheme_table.add_row(anodes=[0], cathodes=[1])
bipolar_scheme_table.add_row(anodes=[0, 1], cathodes=[2, 3])
bipolar_scheme_table.add_row(anodes=[0, 1], cathodes=[2])
bipolar_scheme_table.anodes.table = nwbfile.electrodes
bipolar_scheme_table.cathodes.table = nwbfile.electrodes
bipolar_scheme_region = DynamicTableRegion(
name='electrodes',
data=np.arange(0, 3),
description='desc',
table=bipolar_scheme_table)
ec_series = ElectricalSeries(name='test_ec_series',
description='desc',
data=np.random.rand(100, 3),
rate=1000.,
electrodes=bipolar_scheme_region)
nwbfile.add_acquisition(ec_series)
ecephys_ext = EcephysExt(name='ecephys_ext')
ecephys_ext.bipolar_scheme_table = bipolar_scheme_table
nwbfile.add_lab_meta_data(ecephys_ext)
with NWBHDF5IO('test_nwb.nwb', 'w') as io:
io.write(nwbfile)
with NWBHDF5IO('test_nwb.nwb', 'r', load_namespaces=True) as io:
nwbfile = io.read()
print(nwbfile.acquisition['test_ec_series'].electrodes.table['anodes'][2]['x'])
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
ndx-bipolar-scheme-0.3.1.tar.gz
(13.3 kB
view details)
Built Distribution
File details
Details for the file ndx-bipolar-scheme-0.3.1.tar.gz.
File metadata
- Download URL: ndx-bipolar-scheme-0.3.1.tar.gz
- Upload date:
- Size: 13.3 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.23.0 setuptools/46.1.3.post20200330 requests-toolbelt/0.9.1 tqdm/4.46.0 CPython/3.7.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 3bb26bcb4af8896dd4709376f39b0b4bc62fce85546cde54ab3180d66beae67f |
|
| MD5 | 6da447b8967a201fbe7e58e1753dcfec |
|
| BLAKE2b-256 | 3a71c9b228654e888aa081d1f7a2a867dacb50029930b02d6952ba2f9c9756e5 |
File details
Details for the file ndx_bipolar_scheme-0.3.1-py2.py3-none-any.whl.
File metadata
- Download URL: ndx_bipolar_scheme-0.3.1-py2.py3-none-any.whl
- Upload date:
- Size: 6.4 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.1.1 pkginfo/1.5.0.1 requests/2.23.0 setuptools/46.1.3.post20200330 requests-toolbelt/0.9.1 tqdm/4.46.0 CPython/3.7.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 21a35a3e19529adf9ffaf9f48db42a4a89dc23111301896707bc2eab4ee93f99 |
|
| MD5 | 8795c7f253ca12fc43942c51f736c7b2 |
|
| BLAKE2b-256 | 5456f70a8a3ac39a4c366d0951d02975dfe05961a802b5a368b9cb485e2ccf2e |