extension for fiber photometry data
Project description
ndx-photometry Extension for NWB
Introduction
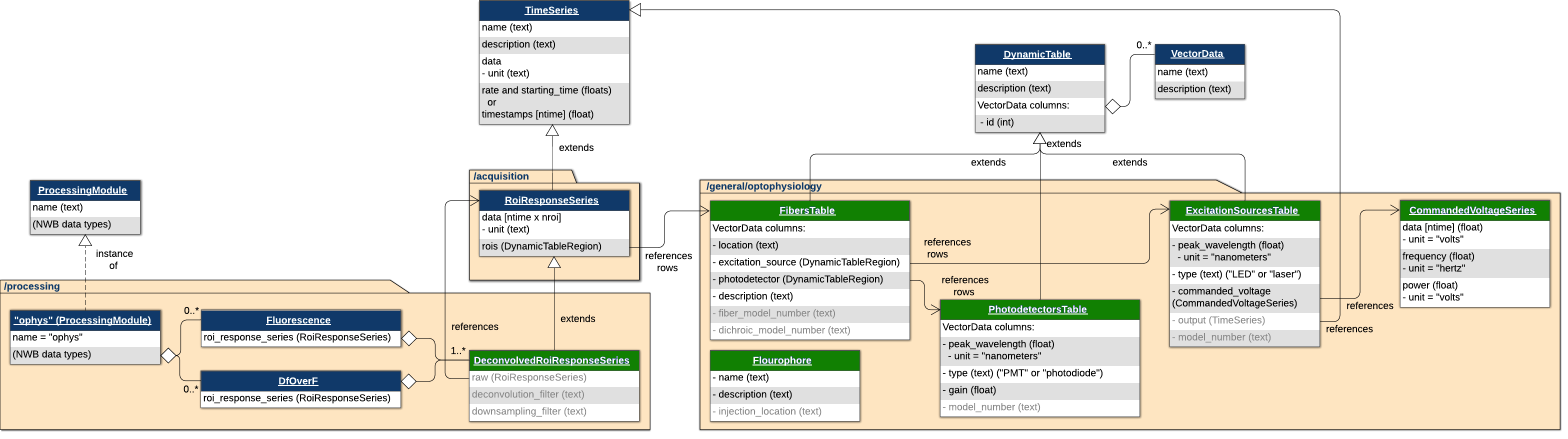
This is an NWB extension for storing photometry recordings and associated metadata. This extension stores photometry information across three folders in the NWB file: acquisition, processing, and general. The acquisiton folder contains an ROIResponseSeries (inherited from pynwb.ophys), which references rows of a FibersTable rather than 2 Photon ROIs. The new types for this extension are in metadata and processing
Metadata
FibersTablestores rows for each fiber with information about the location, excitation, source, photodetector, fluorophore, and more (associated with each fiber).ExcitationSourcesTablestores rows for each excitation source with information about the peak wavelength, source type, and the commanded voltage series of typeCommandedVoltageSeriesPhotodectorsTablestores rows for each photodetector with information about the peak wavelength, type, etc.FluorophoresTablestores rows for each fluorophore with information about the fluorophore itself and the injeciton site.
Processing
DeconvoledROIResponseSeriesstores DfOverF and Fluorescence traces and extendsROIResponseSeriesto contain information about the deconvolutional and downsampling procedures performed.
This extension was developed by Akshay Jaggi, Ben Dichter, and Ryan Ly.
Installation
pip install ndx-photometry
Usage
import datetime
import numpy as np
from pynwb import NWBHDF5IO, NWBFile
from pynwb.core import DynamicTableRegion
from pynwb.ophys import RoiResponseSeries
from ndx_photometry import (
FibersTable,
PhotodetectorsTable,
ExcitationSourcesTable,
DeconvolvedRoiResponseSeries,
MultiCommandedVoltage,
FiberPhotometry,
FluorophoresTable
)
nwbfile = NWBFile(
session_description="session_description",
identifier="identifier",
session_start_time=datetime.datetime.now(datetime.timezone.utc),
)
# In the next ten calls or so, we'll set up the metadata from the bottom of the metadata tree up
# You can follow along here:
# Create a commanded voltage container, this can store one or more commanded voltage series
multi_commanded_voltage = MultiCommandedVoltage(
name="MyMultiCommandedVoltage",
)
# Add a commanded voltage series to this container
commandedvoltage_series = (
multi_commanded_voltage.create_commanded_voltage_series(
name="commanded_voltage",
data=[1.0, 2.0, 3.0],
frequency=30.0,
power=500.0,
rate=30.0,
unit="volts",
)
)
# Create an excitation sources table
excitationsources_table = ExcitationSourcesTable(
description="excitation sources table"
)
# Add one row to the table per excitation source
# You can repeat this in a for-loop for many sources
excitationsources_table.add_row(
peak_wavelength=700.0,
source_type="laser",
commanded_voltage=commandedvoltage_series,
)
photodetectors_table = PhotodetectorsTable(
description="photodetectors table"
)
# Add one row to the table per photodetector
photodetectors_table.add_row(
peak_wavelength=500.0,
type="PMT",
gain=100.0
)
fluorophores_table = FluorophoresTable(
description='fluorophores'
)
fluorophores_table.add_row(
label='dlight',
location='VTA',
coordinates=(3.0,2.0,1.0)
)
fibers_table = FibersTable(
description="fibers table"
)
# Here we add the metadata tables to the metadata section
nwbfile.add_lab_meta_data(
FiberPhotometry(
fibers=fibers_table,
excitation_sources=excitationsources_table,
photodetectors=photodetectors_table,
fluorophores=fluorophores_table
)
)
# Important: we add the fibers to the fibers table _after_ adding the metadata
# This ensures that we can find this data in their tables of origin
fibers_table.add_fiber(
excitation_source=0, #integers indicated rows of excitation sources table
photodetector=0,
fluorophores=[0], #potentially multiple fluorophores, so list of indices
location='my location',
notes='notes'
)
# Here we set up a list of fibers that our recording came from
fibers_ref = DynamicTableRegion(
name="rois",
data=[0], # potentially multiple fibers
description="source fibers",
table=fibers_table
)
# Create a raw roiresponseseries, this is your main acquisition
roi_response_series = RoiResponseSeries(
name="roi_response_series",
description="my roi response series",
data=np.random.randn(100, 1),
unit='F',
rate=30.0,
rois=fibers_ref,
)
# This is your processed data
deconv_roi_response_series = DeconvolvedRoiResponseSeries(
name="DeconvolvedRoiResponseSeries",
description="my roi response series",
data=np.random.randn(100, 1),
unit='F',
rate=30.0,
rois=fibers_ref,
raw=roi_response_series,
)
ophys_module = nwbfile.create_processing_module(
name="ophys", description="fiber photometry"
)
ophys_module.add(multi_commanded_voltage)
nwbfile.add_acquisition(roi_response_series)
ophys_module.add(deconv_roi_response_series)
# write nwb file
filename = 'test.nwb'
with NWBHDF5IO(filename, 'w') as io:
io.write(nwbfile)
# read nwb file and check its contents
with NWBHDF5IO(filename, 'r', load_namespaces=True) as io:
nwbfile = io.read()
# Access and print information about the acquisition
print(nwbfile.acquisition["roi_response_series"])
# Access and print information about the processed data
print(nwbfile.processing['ophys']["DeconvolvedRoiResponseSeries"])
# Access and print all of the metadata
print(nwbfile.lab_meta_data)
This extension was created using ndx-template.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distributions
Built Distribution
File details
Details for the file ndx_photometry-0.2.0-py2.py3-none-any.whl.
File metadata
- Download URL: ndx_photometry-0.2.0-py2.py3-none-any.whl
- Upload date:
- Size: 11.2 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.2 CPython/3.9.12
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | c3c99d4766e97ed5e4235b4e47f0c9d52979a940e6c139643e2ad00d85fc1001 |
|
| MD5 | ac90c92b6bab82762ad542834fa647f1 |
|
| BLAKE2b-256 | 3a1973b46a7447c8345372c5c6790a29641cf31309214eeb8a02295b61ae981e |