NetworkX for ontologies
Project description
NetworkX-based Python library for representing ontologies
Summary
nxontology is a Python library for representing ontologies using a NetworkX graph. Currently, the main area of functionality is computing similarity measures between pairs of nodes.
Usage
Here, we'll use the example metals ontology:
Note that NXOntology represents the ontology as a networkx.DiGraph, where edge direction goes from superterm to subterm.
Given an NXOntology instance, here how to compute intrinsic similarity metrics.
from nxontology.examples import create_metal_nxo
metals = create_metal_nxo()
# Freezing the ontology prevents adding or removing nodes or edges.
# Frozen ontologies cache expensive computations.
metals.freeze()
# Get object for computing similarity, using the Sanchez et al metric for information content.
similarity = metals.similarity("gold", "silver", ic_metric="intrinsic_ic_sanchez")
# Access a single similarity metric
similarity.lin
# Access all similarity metrics
similarity.results()
The final line outputs a dictionary like:
{
'node_0': 'gold',
'node_1': 'silver',
'node_0_subsumes_1': False,
'node_1_subsumes_0': False,
'n_common_ancestors': 3,
'n_union_ancestors': 5,
'batet': 0.6,
'batet_log': 0.5693234419266069,
'ic_metric': 'intrinsic_ic_sanchez',
'mica': 'coinage',
'resnik': 0.8754687373538999,
'resnik_scaled': 0.48860840553061435,
'lin': 0.5581154235118403,
'jiang': 0.41905978419640516,
'jiang_seco': 0.6131471927654584,
}
It's also possible to visualize the similarity between two nodes like:
from nxontology.viz import create_similarity_graphviz
gviz = create_similarity_graphviz(
# similarity instance from above
similarity,
# show all nodes (defaults to union of ancestors)
nodes=list(metals.graph),
)
# draw to PNG file
gviz.draw("metals-sim-gold-silver-all.png"))
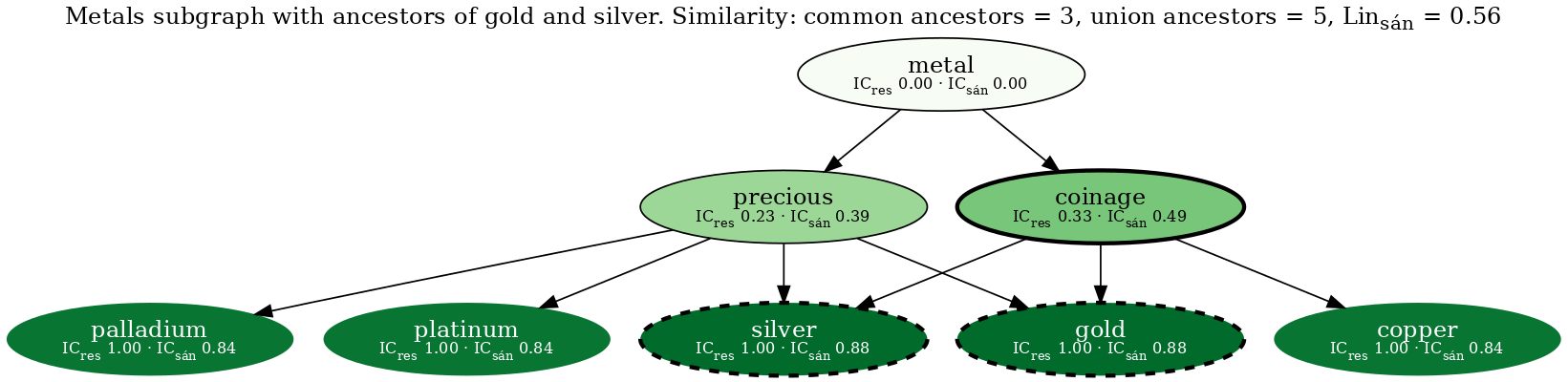
Resulting in the following figure:
The two query nodes (gold & silver) are outlined with a bold dashed line. Node fill color corresponds to the Sánchez information content, such that darker nodes have higher IC. The most informative common ancestor (coinage) is outlined with a bold solid line. Nodes that are not an ancestor of gold or silver have an invisible outline.
Loading ontologies
Pronto supports reading ontologies from the following file formats:
- Open Biomedical Ontologies 1.4:
.oboextension, uses the fastobo parser. - OBO Graphs JSON:
.jsonextension, uses the fastobo parser. - Ontology Web Language 2 RDF/XML:
.owlextension, uses the prontoRdfXMLParser.
The files can be local or at a network location (URL starting with https, http, or ftp). Pronto detects and handles gzip, bzip2, and xz compression.
Here are examples operations on the Gene Ontology, using pronto to load the ontology:
>>> from nxontology.imports import from_file
>>> # versioned URL for the Gene Ontology
>>> url = "http://release.geneontology.org/2021-02-01/ontology/go-basic.json.gz"
>>> nxo = from_file(url)
>>> nxo.n_nodes
44085
>>> # similarity between "myelination" and "neurogenesis"
>>> sim = nxo.similarity("GO:0042552", "GO:0022008")
>>> round(sim.lin, 2)
0.21
>>> import networkx as nx
>>> # Gene Ontology domains are disconnected, expect 3 components
>>> nx.number_weakly_connected_components(nxo.graph)
3
>>> # Note however that the default from_file reader only uses "is a" relationships.
>>> # We can preserve all GO relationship types as follows
>>> from collections import Counter
>>> import pronto
>>> from nxontology import NXOntology
>>> from nxontology.imports import pronto_to_multidigraph, multidigraph_to_digraph
>>> go_pronto = pronto.Ontology(handle=url)
>>> go_multidigraph = pronto_to_multidigraph(go_pronto)
>>> Counter(key for _, _, key in go_multidigraph.edges(keys=True))
Counter({'is a': 71509,
'part of': 7187,
'regulates': 3216,
'negatively regulates': 2768,
'positively regulates': 2756})
>>> go_digraph = multidigraph_to_digraph(go_multidigraph, reduce=True)
>>> go_nxo = NXOntology(go_digraph)
>>> # Notice the similarity increases due to the full set of edges
>>> round(go_nxo.similarity("GO:0042552", "GO:0022008").lin, 3)
0.699
>>> # Note that there is also a dedicated reader for the Gene Ontology
>>> from nxontology.imports import read_gene_ontology
>>> read_gene_ontology(release="2021-02-01")
Users can also create their own networkx.DiGraph to use this package.
Installation
nxontology can be installed with pip from PyPI like:
# standard installation
pip install nxontology
# installation with viz extras
pip install nxontology[viz]
The extra viz dependencies are required for the nxontology.viz module.
This includes pygraphviz, which requires a pre-existing graphviz installation.
Development
Some helpful development commands:
# create a virtual environment for development
python3 -m venv .venv
# activate virtual environment
source .venv/bin/activate
# install package for development
pip install --editable ".[dev,viz]"
# Set up the git pre-commit hooks.
# `git commit` will now trigger automatic checks including linting.
pre-commit install
# Run all pre-commit checks (CI will also run this).
pre-commit run --all
# run tests
pytest
Releases are created on GitHub.
The release action defined by release.yaml will build the distribution and upload to PyPI.
The package version is automatically generated from the git tag by setuptools_scm.
Bibliography
Here's a list of alternative projects with code for computing semantic similarity measures on ontologies:
- Semantic Measures Library & ToolKit at sharispe/slib in Java.
- DiShIn at lasigeBioTM/DiShIn in Python.
- Sematch at gsi-upm/sematch in Python.
- ontologySimilarity mirrored at cran/ontologySimilarity. Part of the ontologyX suite of R packages.
- Materials for Machine Learning with Ontologies at bio-ontology-research-group/machine-learning-with-ontologies (compilation)
Below are a list of references related to ontology-derived measures of similarity.
Feel free to add any reference that provides useful context and details for algorithms supported by this package.
Metadata for a reference can be generated like manubot cite --yml doi:10.1016/j.jbi.2011.03.013.
Adding CSL YAML output to media/bibliography.yaml will cache the metadata and allow manual edits in case of errors.
-
Semantic Similarity in Biomedical Ontologies
Catia Pesquita, Daniel Faria, André O. Falcão, Phillip Lord, Francisco M. Couto
PLoS Computational Biology (2009-07-31) https://doi.org/cx8h87
DOI: 10.1371/journal.pcbi.1000443 · PMID: 19649320 · PMCID: PMC2712090 -
An Intrinsic Information Content Metric for Semantic Similarity in WordNet.
Nuno Seco, Tony Veale, Jer Hayes
In Proceedings of the 16th European Conference on Artificial Intelligence (ECAI-04), (2004) https://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.1065.1695 -
Metrics for GO based protein semantic similarity: a systematic evaluation
Catia Pesquita, Daniel Faria, Hugo Bastos, António EN Ferreira, André O Falcão, Francisco M Couto
BMC Bioinformatics (2008-04-29) https://doi.org/cmcgw6
DOI: 10.1186/1471-2105-9-s5-s4 · PMID: 18460186 · PMCID: PMC2367622 -
Semantic similarity and machine learning with ontologies
Maxat Kulmanov, Fatima Zohra Smaili, Xin Gao, Robert Hoehndorf
Briefings in Bioinformatics (2020-10-13) https://doi.org/ghfqkt
DOI: 10.1093/bib/bbaa199 · PMID: 33049044 -
Semantic Similarity in a Taxonomy: An Information-Based Measure and its Application to Problems of Ambiguity in Natural Language
P. Resnik
Journal of Artificial Intelligence Research (1999-07-01) https://doi.org/gftcpz
DOI: 10.1613/jair.514 -
An Information-Theoretic Definition of Similarity
Dekang Lin
ICML (1998) https://api.semanticscholar.org/CorpusID:5659557 -
ontologyX: a suite of R packages for working with ontological data
Daniel Greene, Sylvia Richardson, Ernest Turro
Bioinformatics (2017-01-05) https://doi.org/f9k7sx
DOI: 10.1093/bioinformatics/btw763 · PMID: 28062448 · PMCID: PMC5386138 -
Metric of intrinsic information content for measuring semantic similarity in an ontology
Md. Hanif Seddiqui, Masaki Aono
Proceedings of the Seventh Asia-Pacific Conference on Conceptual Modelling - Volume 110 (2010-01-01) https://dl.acm.org/doi/10.5555/1862330.1862343
ISBN: 9781920682927 -
Disjunctive shared information between ontology concepts: application to Gene Ontology
Francisco M Couto, Mário J Silva
Journal of Biomedical Semantics (2011) https://doi.org/fnb73v
DOI: 10.1186/2041-1480-2-5 · PMID: 21884591 · PMCID: PMC3200982 -
A framework for unifying ontology-based semantic similarity measures: A study in the biomedical domain
Sébastien Harispe, David Sánchez, Sylvie Ranwez, Stefan Janaqi, Jacky Montmain
Journal of Biomedical Informatics (2014-04) https://doi.org/f52557
DOI: 10.1016/j.jbi.2013.11.006 · PMID: 24269894 -
Semantic Similarity in Cheminformatics
João D. Ferreira, Francisco M. Couto
IntechOpen (2020-07-15) https://doi.org/ghh2d4
DOI: 10.5772/intechopen.89032 -
An ontology-based measure to compute semantic similarity in biomedicine
Montserrat Batet, David Sánchez, Aida Valls
Journal of Biomedical Informatics (2011-02) https://doi.org/dfhkjv
DOI: 10.1016/j.jbi.2010.09.002 · PMID: 20837160 -
Semantic similarity in the biomedical domain: an evaluation across knowledge sources
Vijay N Garla, Cynthia Brandt
BMC Bioinformatics (2012-10-10) https://doi.org/gb8vpn
DOI: 10.1186/1471-2105-13-261 · PMID: 23046094 · PMCID: PMC3533586 -
Semantic similarity estimation in the biomedical domain: An ontology-based information-theoretic perspective
David Sánchez, Montserrat Batet
Journal of Biomedical Informatics (2011-10) https://doi.org/d2436q
DOI: 10.1016/j.jbi.2011.03.013 · PMID: 21463704 -
Ontology-based information content computation
David Sánchez, Montserrat Batet, David Isern
Knowledge-Based Systems (2011-03) https://doi.org/cwzw4r
DOI: 10.1016/j.knosys.2010.10.001 -
Leveraging synonymy and polysemy to improve semantic similarity assessments based on intrinsic information content
Montserrat Batet, David Sánchez
Artificial Intelligence Review (2019-06-03) https://doi.org/ghnfmt
DOI: 10.1007/s10462-019-09725-4 -
An intrinsic information content-based semantic similarity measure considering the disjoint common subsumers of concepts of an ontology
Abhijit Adhikari, Biswanath Dutta, Animesh Dutta, Deepjyoti Mondal, Shivang Singh
Journal of the Association for Information Science and Technology (2018-08) https://doi.org/gd2j5b
DOI: 10.1002/asi.24021
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file nxontology-0.3.3.tar.gz.
File metadata
- Download URL: nxontology-0.3.3.tar.gz
- Upload date:
- Size: 132.2 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.7.0 importlib_metadata/4.8.2 pkginfo/1.8.2 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.8.12
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 13a795c409d2d91b36c75a3a296310f4db12497b9ac648020bfcb5f54e3af514 |
|
| MD5 | 1cb25395211e07572de6f598a42a5008 |
|
| BLAKE2b-256 | 6cdfc6e3c318fb294cab27f61d9d1cf66bed60033473f9317c65577652c15f3a |
File details
Details for the file nxontology-0.3.3-py3-none-any.whl.
File metadata
- Download URL: nxontology-0.3.3-py3-none-any.whl
- Upload date:
- Size: 33.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.7.0 importlib_metadata/4.8.2 pkginfo/1.8.2 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.8.12
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 377f13d014b238c6a03f26b40525343c312a8379424d0f1be4e98f2bdcd629c2 |
|
| MD5 | b7e64ef68212736b0c626c375459e9bb |
|
| BLAKE2b-256 | bc27d67aff2ff53d890e5e7b3dc75be5dc2d6ace9669827659ebf955267f8599 |