MAPR is a Python plugin for OMERO.web
Project description


MAPR
OMERO.mapr is an OMERO.web app that enables browsing of data through attributes linked to images in the form of Map Annotations.
It is used extensively by the Image Data Resource, allowing users to find data by various categories such as Genes, Phenotypes, Organism etc.
In OMERO, Map Annotations are lists of named attributes or “Key-Value Pairs” that can be used to annotate many types of data. Annotations can be assigned a namespace to indicate the origin and purpose of the annotation.
Map Annotations created by users via the Insight client or webclient all have the namespace openmicroscopy.org/omero/client/mapAnnotation, whereas other Map Annotations created via the OMERO API by other tools should have their own distinct namespace.
We can configure OMERO.mapr to search for Map Annotations of specified namespace, looking for Values under specifed Keys. For example, seach for values under key Gene Symbol or Gene Identifier and namespace openmicroscopy.org/mapr/gene.
Requirements
OMERO.web 5.6 or newer.
Installing from Pypi
This section assumes that an OMERO.web is already installed. NB: Configuration of the settings (see below) is not optional and is required for the app to work.
Install the app using pip:
$ pip install omero-mapr
Add the app to the list of installed apps:
$ omero config append omero.web.apps '"omero_mapr"'
Config Settings
You need to configure the namespaces and keys that you want users to be able to search for.
User-edited Map Annotations
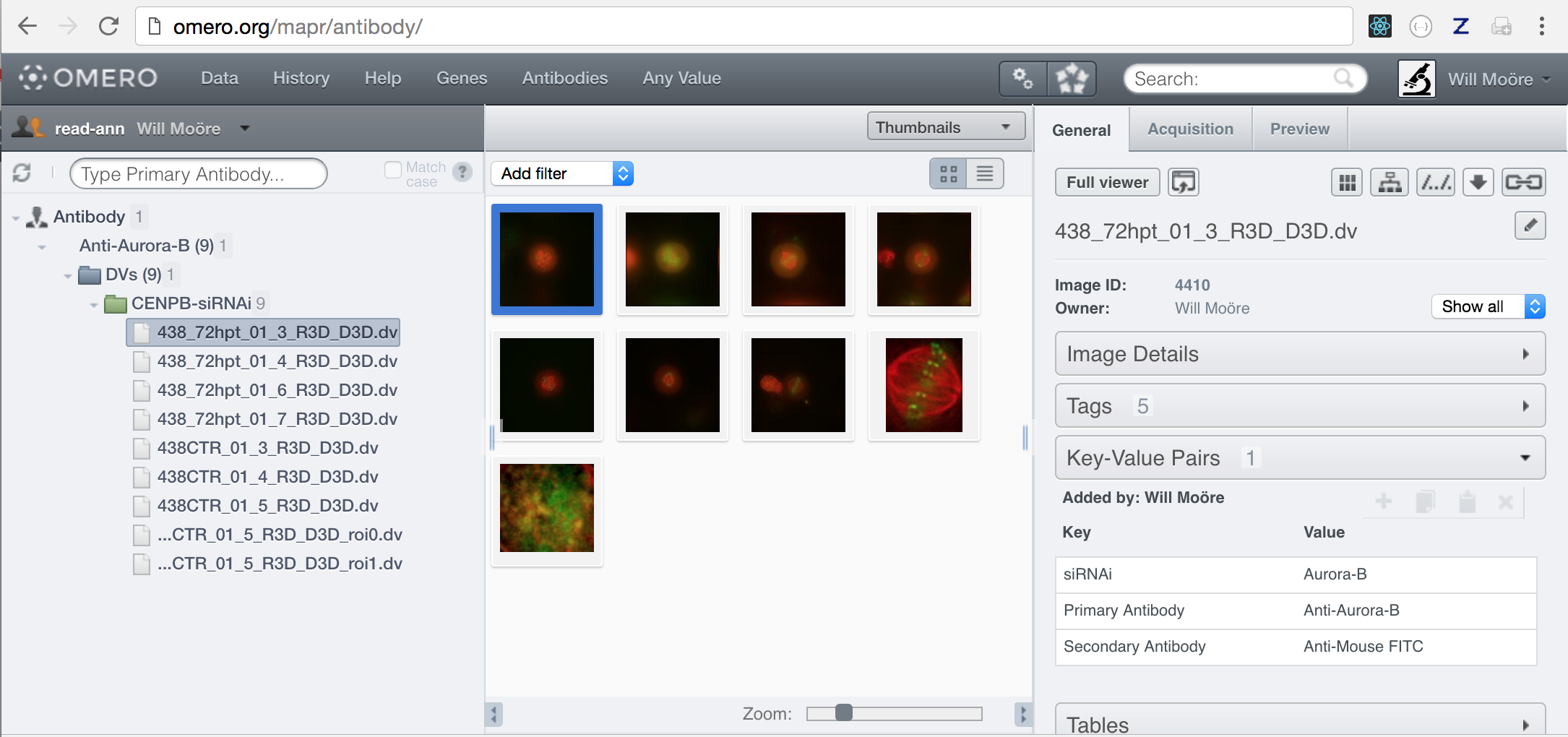
Map Annotations that are added using the webclient or Insight in the Key-Value panel use a specific “client” namespace. We can therefore configure OMERO.mapr to search for these annotations using the namespace openmicroscopy.org/omero/client/mapAnnotation. For example, to search for “Primary Antibody” or “Secondary Antibody” values, we can add:
$ omero config append omero.web.mapr.config '{"menu": "antibody", "config":{"default":["Primary Antibody"], "all":["Primary Antibody", "Secondary Antibody"], "ns":["openmicroscopy.org/omero/client/mapAnnotation"], "label":"Antibody"}}'
We can add an “Antibodies” link to the top of the webclient page to take us to the Antibodies search page. The link tooltip is “Find Antibody values”. The viewname should be in the form maprindex_{menu} where {menu} is the the menu value in the previous config.
$ omero config append omero.web.ui.top_links '["Antibodies", {"viewname": "maprindex_antibody"}, {"title": "Find Antibody values"}]'
After restarting web, we can now search for Antibodies:
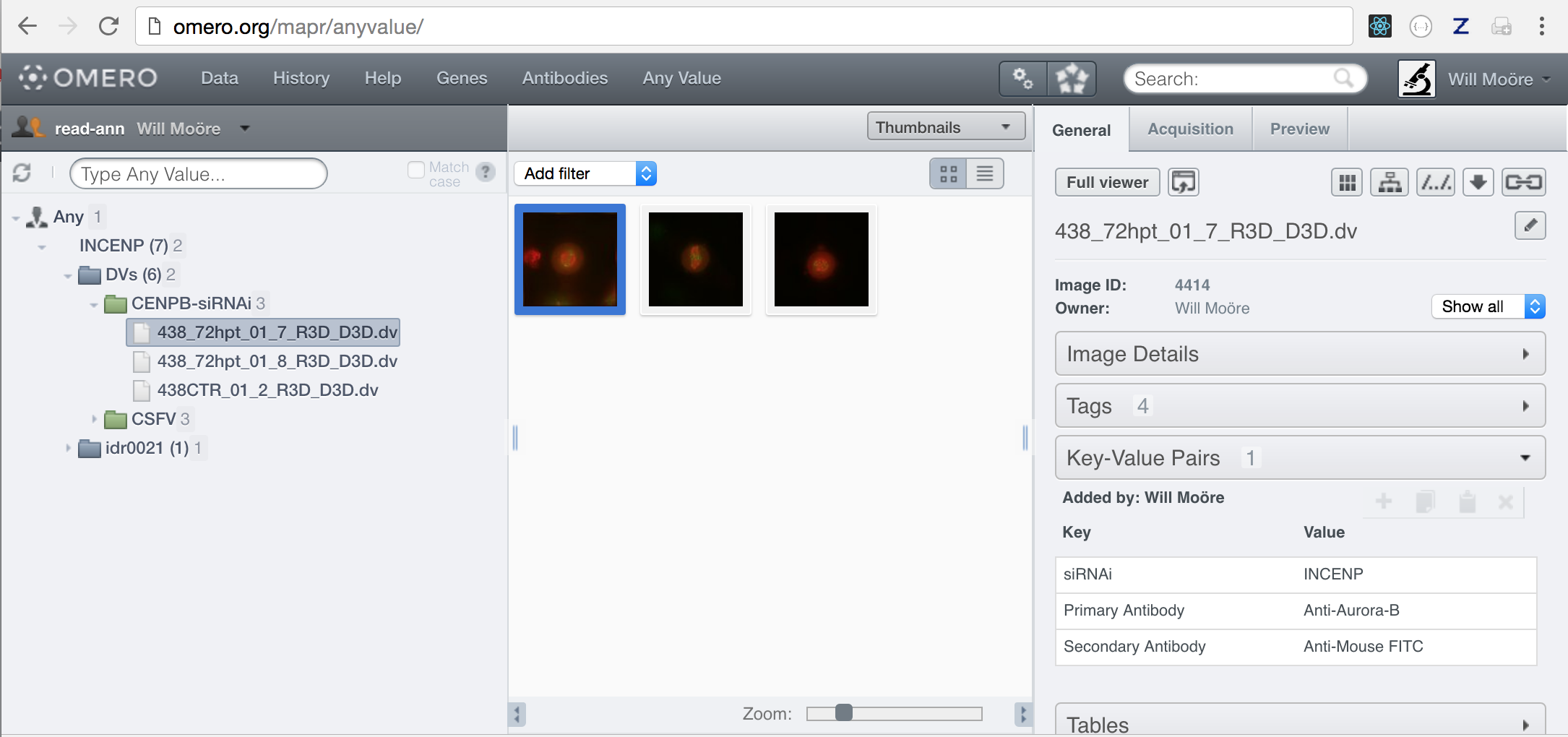
We can also specify an empty list of keys to search for any value.
$ omero config append omero.web.mapr.config '{"menu": "anyvalue", "config":{"default":["Any Value"], "all":[], "ns":["openmicroscopy.org/omero/client/mapAnnotation"], "label":"Any"}}'
# Top link
$ omero config append omero.web.ui.top_links '["Any Value", {"viewname": "maprindex_anyvalue"}, {"title": "Find Any Value"}]'
After restarting web, we can now search for any Value, such as “INCENP”:
Other Map Annotations
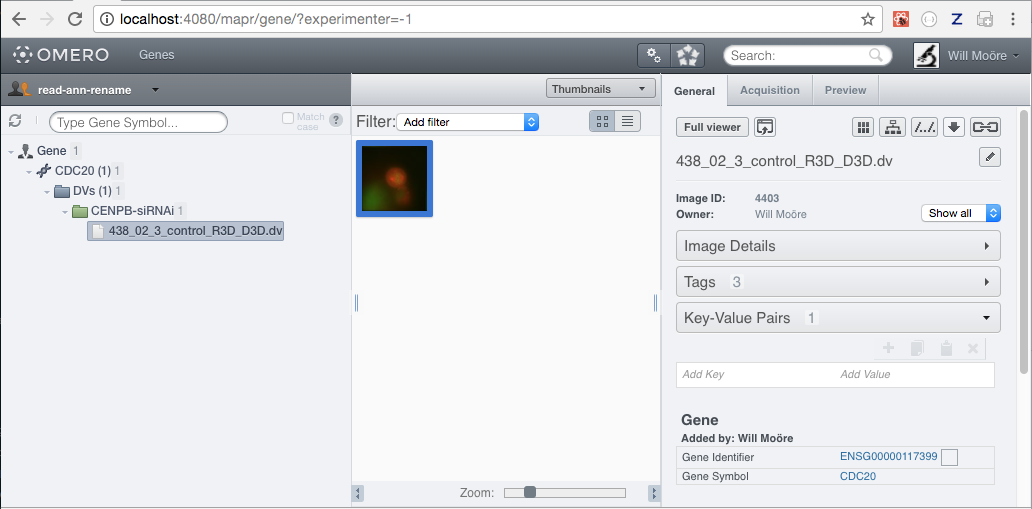
In this example we want to search for Map Annotations of namespace openmicroscopy.org/mapr/gene searching for attributes under the Gene Symbol and Gene Identifier keys.
$ omero config append omero.web.mapr.config '{"menu": "gene","config": {"default": ["Gene Symbol"],"all": ["Gene Symbol", "Gene Identifier"],"ns": ["openmicroscopy.org/mapr/gene"],"label": "Gene"}}'
Now add a top link of Genes with tooltip Find Gene annotations that will take us to the gene search page. The query_string parameters are added to the URL, with "experimenter": -1 specifying that we want to search across all users.
$ omero config append omero.web.ui.top_links '["Genes", {"viewname": "maprindex_gene", "query_string": {"experimenter": -1}}, {"title": "Find Gene annotations"}]'
Finally, we can add a map annotation to an Image that is in a Screen -> Plate -> Well or Project -> Dataset -> Image hierarchy. This code uses the OMERO Python API to add a map annotation corresponding to the configuration above:
key_value_data = [["Gene Identifier","ENSG00000117399"],
["Gene Identifier URL", "http://www.ensembl.org/id/ENSG00000117399"],
["Gene Symbol","CDC20"]]
map_ann = omero.gateway.MapAnnotationWrapper(conn)
map_ann.setValue(key_value_data)
map_ann.setNs("openmicroscopy.org/mapr/gene")
map_ann.save()
image = conn.getObject('Image', 2917)
image.linkAnnotation(map_ann)
Now restart OMERO.web as normal for the configuration above to take effect. You should now be able to browse to a Genes page and search for CDC20 or ENSG00000117399.
External URL Favicons
Mapr can automatically convert URLs into favicon links. To use this feature the key such as Gene Identifier must be in the “all” list of a config as shown above and the Gene Identifier key-value pair must be followed by a key-value pair called Gene Identifier URL. A favicon linked to the external URL will be appended to the Gene Identifier row, and the Gene Identifier URL key-value pair will be hidden. OMERO.web must be configured with the Django redis cache https://docs.openmicroscopy.org/omero/5/sysadmins/unix/install-web/walkthrough/omeroweb-install-centos7-ice3.6.html?highlight=redis#configuring-omero-web which is used to cache the favicons that are obtained using a Google service.
Testing
Testing MAPR requires OMERO.server running. Run tests (includes self-contained OMERO.server, requires docker):
docker-compose -f docker/docker-compose.yml up --build --abort-on-container-exit docker-compose -f docker/docker-compose.yml rm -fv
License
MAPR is released under the AGPL.
Copyright
2016-2021, The Open Microscopy Environment
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file omero-mapr-0.5.0.tar.gz.
File metadata
- Download URL: omero-mapr-0.5.0.tar.gz
- Upload date:
- Size: 54.8 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.8.0 pkginfo/1.8.2 readme-renderer/34.0 requests/2.27.1 requests-toolbelt/0.9.1 urllib3/1.26.9 tqdm/4.63.0 importlib-metadata/4.11.3 keyring/23.5.0 rfc3986/2.0.0 colorama/0.4.4 CPython/3.8.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 68d0b578aba49eeb68124aa979ea5c1edefe3db3648b9088436a44564a49b4d5 |
|
| MD5 | 443f648a0585dc80fda72a631a3dbce0 |
|
| BLAKE2b-256 | da5e8a7854100b8b62defce33d34dbceca92a8319b7b161a606131031777102e |
File details
Details for the file omero_mapr-0.5.0-py3-none-any.whl.
File metadata
- Download URL: omero_mapr-0.5.0-py3-none-any.whl
- Upload date:
- Size: 66.5 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.8.0 pkginfo/1.8.2 readme-renderer/34.0 requests/2.27.1 requests-toolbelt/0.9.1 urllib3/1.26.9 tqdm/4.63.0 importlib-metadata/4.11.3 keyring/23.5.0 rfc3986/2.0.0 colorama/0.4.4 CPython/3.8.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 122ffee56ed4065effd523d2518339af9d49220291c925731e3fc4f484ff142f |
|
| MD5 | 039c16a7b8f390b84684df4f427ebf3a |
|
| BLAKE2b-256 | f4b7a417f71ee16827a80f0e1a03081a0e2d83b4df29afb00bbe58f648f3563b |















