A tool to work with pre-computed large pubmed embedding.
Project description
Building PubMed embedding, automatically.
Install the package
As usual, just install from Pypi:
pip install pubmed_embeddingUsage examples
You can retrieve embedding for PubMed IDs of interest as such:
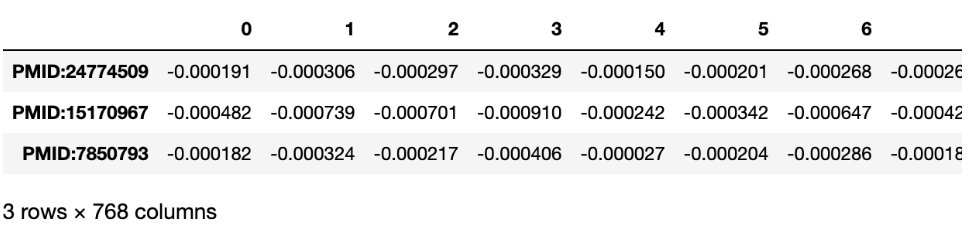
BERT
from pubmed_embedding import get_pubmed_embedding_from_curies
pubmed_ids = ["PMID:24774509", "PMID:15170967", "PMID:7850793"]
bert_features = get_pubmed_embedding_from_curies(
curies=pubmed_ids,
version="pubmed_bert_30_11_2022"
)And the result is:
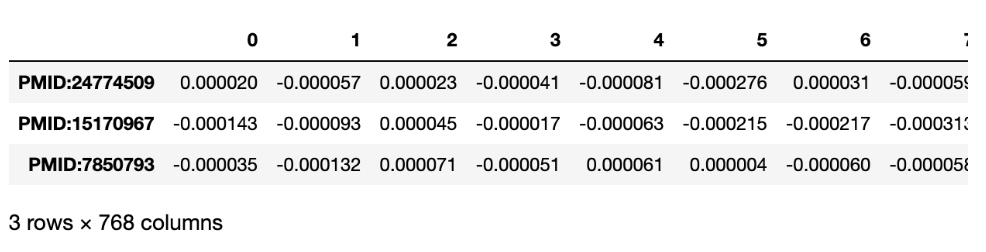
SciBERT
scibert_features = get_pubmed_embedding_from_curies(
curies=pubmed_ids,
version="pubmed_scibert_30_11_2022"
)And the result is:
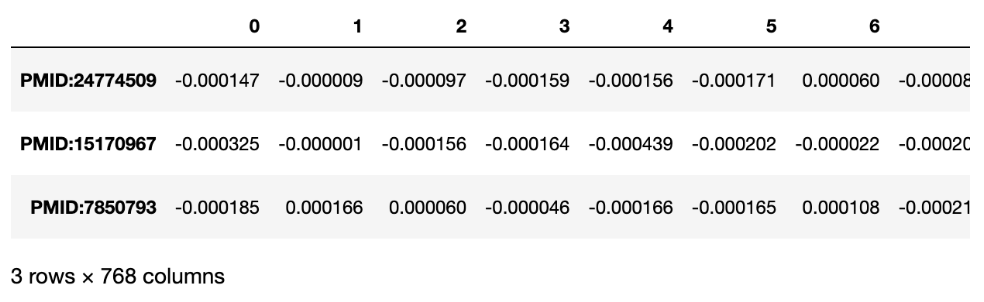
Specter
spected_features = get_pubmed_embedding_from_curies(
curies=pubmed_ids,
version="pubmed_specter_30_11_2022"
)And the result is:
Citing this work
If you have found these datasets useful, please do cite:
@software{cappellettiPubMed2022,
author = {Luca, Cappelletti and Tommaso, Fontana and Justin, Reese},
month = {12},
title = {{BM25-weighted BERT-based embedding of PubMed}},
url = {https://github.com/LucaCappelletti94/pubmed_embedding},
version = {1.0.12},
year = {2022}
}Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
pubmed_embedding-1.0.13.tar.gz
(30.0 kB
view hashes)