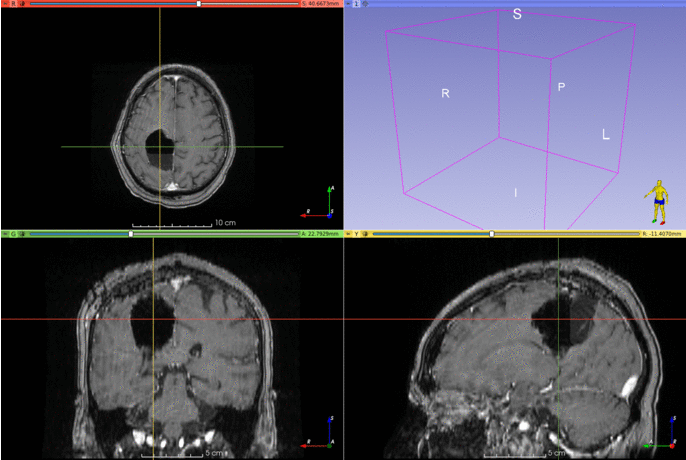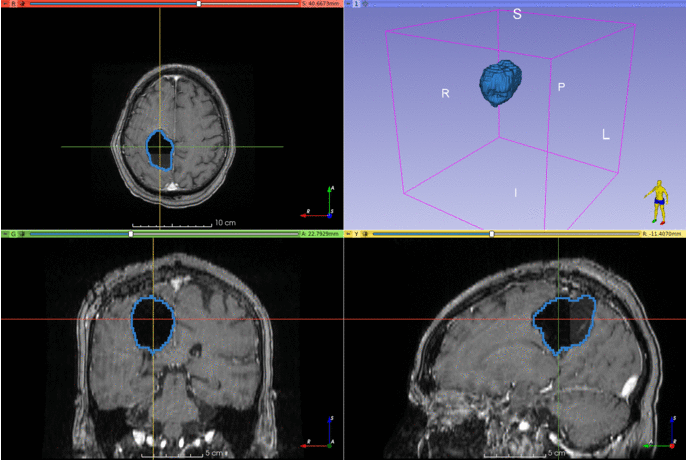
Automatic segmentation of epilepsy neurosurgery resection cavity.
Project description
RESSEG
Automatic segmentation of postoperative brain resection cavities from magnetic resonance images (MRI) using a convolutional neural network (CNN) trained with PyTorch 1.7.1.
Installation
It's recommended to use conda.
A 6-GB GPU is large enough to segment an image in an MNI space of size 193 × 229 × 193.
conda create -n resseg python=3.8 -y
conda activate resseg
pip install light-the-torch
ltt install torch
pip install resseg
resseg --help
Usage
Below are two examples of cavity segmentation for tumor and epilepsy surgery. The epilepsy example includes registration to the MNI space. Both examples can be run online using Google Colab:
BITE
Example using an image from the Brain Images of Tumors for Evaluation database (BITE).
BITE=`resseg-download bite`
resseg $BITE -o bite_seg.nii.gz
EPISURG
Example using an image from the EPISURG dataset.
Segmentation works best when images are in the MNI space, so resseg includes a tool
for this purpose (requires `antspyx).
pip install antspyx
EPISURG=`resseg-download episurg`
resseg-mni $EPISURG -t episurg_to_mni.tfm
resseg $EPISURG -o episurg_seg.nii.gz -t episurg_to_mni.tfm
Trained model
The trained model can be used without installing resseg, but you'll need to install unet first:
pip install unet==0.7.7
Then, in Python:
import torch
repo = 'fepegar/resseg'
model_name = 'ressegnet'
model = torch.hub.load(repo, model_name, pretrained=True)
Graphical user interface using 3D Slicer
There is an experimental graphical user interface (GUI) built on top of 3D Slicer.
Visit this repository for additional information and installation instructions.
Plotting resected structures
A quantitative analysis of the resected structures can be performed using a brain parcellation computed using GIF (3.0) or FreeSurfer.
from resseg.parcellation import GIFParcellation, FreeSurferParcellation
parcellation_path = 't1_seg_gif.nii.gz'
cavity_seg_on_preop_path = 'cavity_on_preop.nii.gz'
parcellation = GIFParcellation(parcellation_path)
I used a sphere near the hippocampus to simulate the resection cavity segmentation, and the GIF parcellation in the FPG dataset of TorchIO.
parcellation.print_percentage_of_resected_structures(cavity_seg_on_preop_path)
Percentage of each resected structure:
100% of Left vessel
83% of Left Inf Lat Vent
59% of Left Amygdala
58% of Left Hippocampus
26% of Left PIns posterior insula
24% of Left PP planum polare
21% of Left Basal Forebrain
18% of Left Claustrum
16% of Left PHG parahippocampal gyrus
15% of Left Pallidum
15% of Left Ent entorhinal area
13% of Left FuG fusiform gyrus
13% of Left Temporal White Matter
11% of Left Putamen
10% of Left Insula White Matter
5% of Left ITG inferior temporal gyrus
5% of Left periventricular white matter
5% of Left Ventral DC
The resection volume is composed of:
30% is Left Temporal White Matter
12% is Left Hippocampus
10% is Left Insula White Matter
7% is Left FuG fusiform gyrus
6% is Left Amygdala
4% is Left ITG inferior temporal gyrus
4% is Left PP planum polare
3% is Left Putamen
3% is Left Claustrum
3% is Left PIns posterior insula
3% is Left PHG parahippocampal gyrus
2% is [Unkown label: 4]
1% is Left Ent entorhinal area
1% is Left Pallidum
1% is Left Inf Lat Vent
1% is Left Ventral DC
parcellation.plot_bars(cavity_seg_on_preop_path)
parcellation.plot_pie(cavity_seg_on_preop_path)
Credit
If you use this library for your research, please cite the following publications:
If you use the EPISURG dataset, which was used to train the model, please cite the following publication:
See also
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file resseg-0.3.7.tar.gz.
File metadata
- Download URL: resseg-0.3.7.tar.gz
- Upload date:
- Size: 13.7 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.8.1 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.2 CPython/3.7.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 9247046a4675ded61a746d0868f73c2d4b779f13f750f9b7aac47e8a8b60bf48 |
|
| MD5 | bd9a4213d2e881d909531be2fa4db454 |
|
| BLAKE2b-256 | 76c01bc9278e46e874ce5cd0921f523f41a1766f3f97f892b3c1f3842269c73a |
File details
Details for the file resseg-0.3.7-py2.py3-none-any.whl.
File metadata
- Download URL: resseg-0.3.7-py2.py3-none-any.whl
- Upload date:
- Size: 15.2 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.8.1 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.2 CPython/3.7.1
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | bde5a6cae01d6e3424d2d0c408b4fea66e3eb1bfc6f917307b55d4b01fb5e4e6 |
|
| MD5 | 8e29d22c8e938b67b4e2ad01e444b687 |
|
| BLAKE2b-256 | 9db228f7e928be2ddb2059d85fd6f74378aab8e714575f1c04425e693f389a99 |