Transfer learning with Architecture Surgery on Single-cell data
Project description
|PyPI| |PyPIDownloads| |Docs| |travis|
scArches - single-cell architecture surgery
.. raw:: html
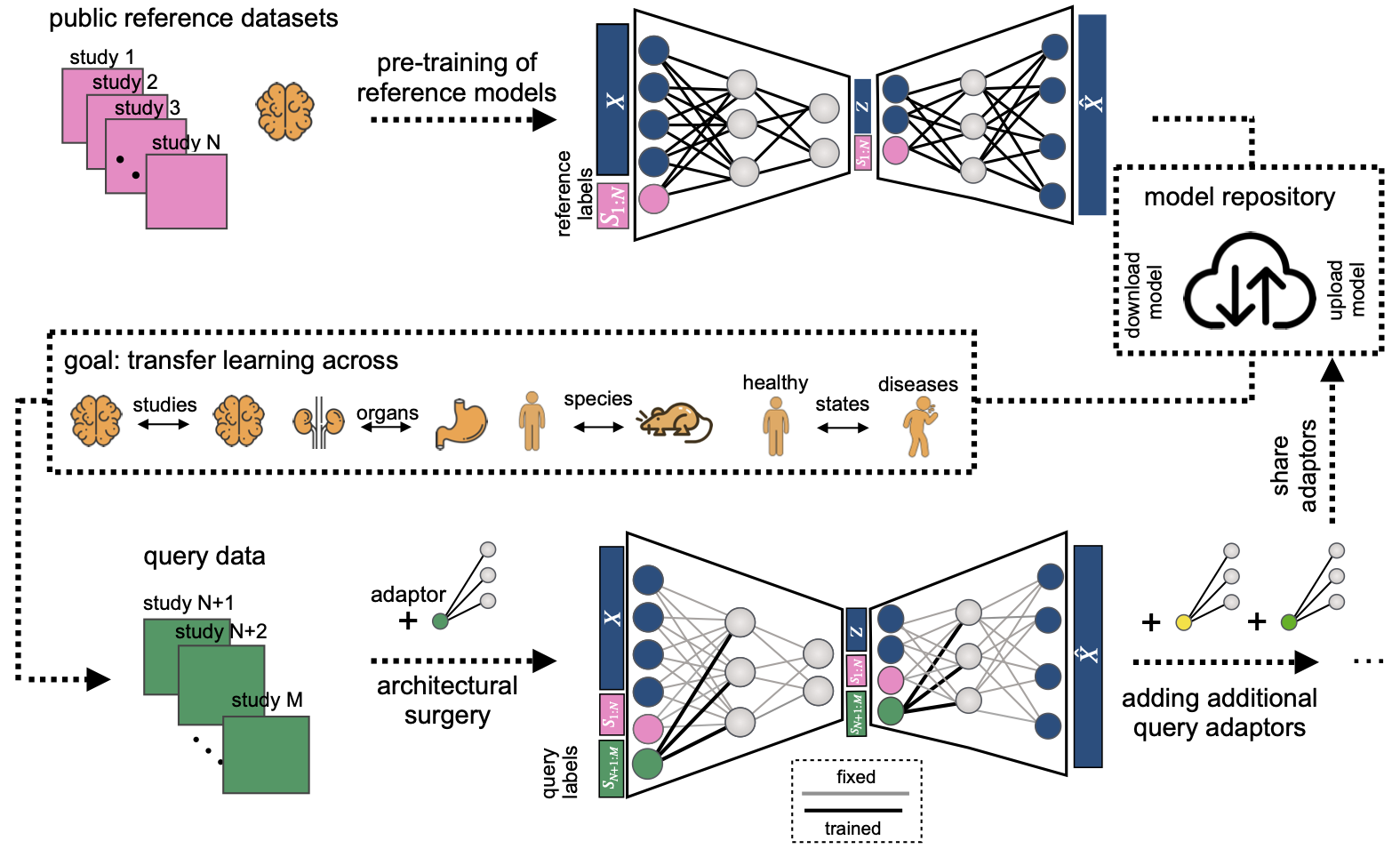
scArches is a package to integrate newly produced single-cell datasets into integrated reference atlases. Our method can facilitate large collaborative projects with decentralized training and integration of multiple datasets by different groups. scArches is compatible with scanpy <https://scanpy.readthedocs.io/en/stable/>_. and hosts efficient implementations of several conditional generative models for single-cell data.
What can you do with scArches?
- Construct single or multi-modal (CITE-seq) reference atlases and share the trained model and the data (if possible).
- Download a pre-trained model for your atlas of interest, update it with new datasets and share with your collaborators.
- Project and integrate query datasets on the top of a reference and use latent representation for downstream tasks, e.g.:diff testing, clustering, classification
What are the different models?
scArches is an algorithm to map to project query on the top of reference datasets and applies to different models. Here we provide a short explanation and hints on when to use which model. Our models are divided into three categories:
Unsupervised
This class of algorithms require no cell type labels, meaning that you can create a reference and project a query without having access to cell type labels.
We implemented two algorithms:
-
scVI (
Lopez et al., 2018 <https://www.nature.com/articles/s41592-018-0229-2>_): Requires access to raw counts values for data integration and assumes count distribution on the data (NB, ZINB, Poisson). -
trVAE (
Lotfollahi et al.,2020 <https://academic.oup.com/bioinformatics/article/36/Supplement_2/i610/6055927?guestAccessKey=71253caa-1779-40e8-8597-c217db539fb5>_): It supports both normalized log transformed or count data as input and applies additional MMD loss to have better merging in the latent space.
Supervised and Semi-supervised
This class of algorithms assumes the user has access to cell type labels when creating the reference data and usually perform better integration compared to. unsupervised methods. However, query data still can be unlabeled. In addition to integration, you can classify your query cells using
these methods.
-
scANVI (
Xu et al., 2019 <https://www.biorxiv.org/content/10.1101/532895v1>_): It needs cell type labels for reference data. Your query data can be either unlabeled or labeled. In the case of unlabeled query data, you can use this method to also classify your query cells using reference labels. -
scGen (
Lotfollahi et al., 2019 <https://www.nature.com/articles/s41592-019-0494-8>_): This method requires cell-type labels for both reference building and query mapping. The query mapping for this method solely relies on the integrated reference and requre no fine-tuning.
Multi-modal These algorithms can be used to construct multi-modal references atlas and map query data from either modality on the top of the reference.
- totalVI (
Gayoso al., 2019 <https://www.biorxiv.org/content/10.1101/532895v1>_): This model can be used to build multi-modal CITE-seq reference atalses. Query datasets can be either from sc-RNAseq or CITE-seq. In addition to integrating query with reference, one can use this model to impute the Proteins in the query datasets.
Usage and installation
See here <https://scarches.readthedocs.io/>_ for documentation and tutorials.
Support and contribute
If you have a question or new architecture or a model that could be integrated into our pipeline, you can
post an issue <https://github.com/theislab/scarches/issues/new>__ or reach us by email <mailto:cottoneyejoe.server@gmail.com,mo.lotfollahi@gmail.com,mohsen.naghipourfar@gmail.com>_.
Reference
If scArches is useful in your research, please consider citing this preprint <https://www.biorxiv.org/content/10.1101/2020.07.16.205997v1/>_.
.. |PyPI| image:: https://img.shields.io/pypi/v/scarches.svg :target: https://pypi-hypernode.com/project/scarches
.. |PyPIDownloads| image:: https://pepy.tech/badge/scarches :target: https://pepy.tech/project/scarches
.. |Docs| image:: https://readthedocs.org/projects/scarches/badge/?version=latest :target: https://scarches.readthedocs.io
.. |travis| image:: https://travis-ci.com/theislab/scarches.svg?branch=master :target: https://travis-ci.com/theislab/scarches
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file scArches-0.4.0.tar.gz.
File metadata
- Download URL: scArches-0.4.0.tar.gz
- Upload date:
- Size: 48.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.6.3 pkginfo/1.7.1 requests/2.22.0 requests-toolbelt/0.9.1 tqdm/4.62.0 CPython/3.8.10
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | abccd6e32a009bc12bf892f1f95d3ccc6a787d56ed3c0acbaad57c92bf611c89 |
|
| MD5 | 8afcb84fe27007d1648e27ad7646b72a |
|
| BLAKE2b-256 | be8f92d1abfde388b99ebd39be6d985531563f9fcb871326255dfee9e1824c83 |
File details
Details for the file scArches-0.4.0-py3-none-any.whl.
File metadata
- Download URL: scArches-0.4.0-py3-none-any.whl
- Upload date:
- Size: 64.4 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.4.2 importlib_metadata/4.6.3 pkginfo/1.7.1 requests/2.22.0 requests-toolbelt/0.9.1 tqdm/4.62.0 CPython/3.8.10
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 44f6a247cf236f9be8f106f966069ef88ef43aeaeead35683c1f4819541a3844 |
|
| MD5 | 771dc3357b521e8418f76f00c836f9fd |
|
| BLAKE2b-256 | c130c254489805952cd7b457bf82a793b4438597fba774037c9567c43a21937a |














