Single-Cell Analysis in Python.
Project description
Scanpy – Single-Cell Analysis in Python
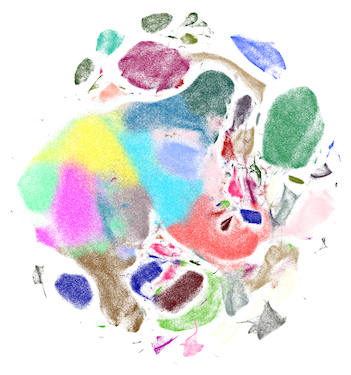
Scanpy is a scalable toolkit for analyzing single-cell gene expression data. It includes preprocessing, visualization, clustering, pseudotime and trajectory inference and differential expression testing. The Python-based implementation efficiently deals with datasets of more than one million cells.
Read the documentation. If Scanpy is useful for your research, consider citing Genome Biology (2018).
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
scanpy-1.3.5.tar.gz
(240.5 kB
view details)
File details
Details for the file scanpy-1.3.5.tar.gz.
File metadata
- Download URL: scanpy-1.3.5.tar.gz
- Upload date:
- Size: 240.5 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/1.12.1 pkginfo/1.4.2 requests/2.18.4 setuptools/39.2.0 requests-toolbelt/0.8.0 tqdm/4.26.0 CPython/3.6.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 22ec896d232c1586fab8bd5a989c0a1251b840f87a67e466a0d784b3b10d0782 |
|
| MD5 | 4853fdd9f37ecb5082097082f28ec695 |
|
| BLAKE2b-256 | f61a83abbd428fbd060b041aa5ccf884a3cbba3c7e21342f5c76107386cc6025 |

















