RNA velocity generalized through dynamical modeling
Project description
scVelo - RNA velocity generalized through dynamical modeling
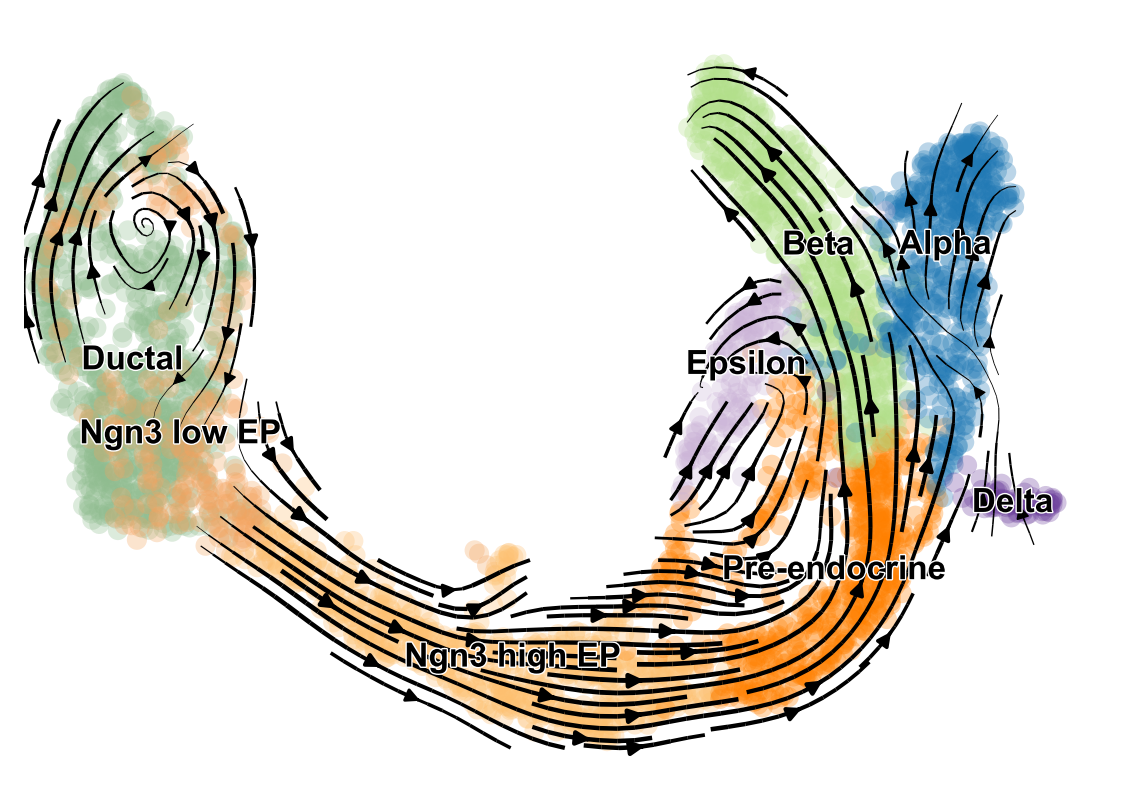
scVelo is a scalable toolkit for RNA velocity analysis in single cells, based on Bergen et al. (Nature Biotech, 2020).
RNA velocity enables the recovery of directed dynamic information by leveraging splicing kinetics. scVelo generalizes the concept of RNA velocity (La Manno et al., Nature, 2018) by relaxing previously made assumptions with a stochastic and a dynamical model that solves the full transcriptional dynamics. It thereby adapts RNA velocity to widely varying specifications such as non-stationary populations.
scVelo is compatible with scanpy and hosts efficient implementations of all RNA velocity models.
scVelo’s key applications
estimate RNA velocity to study cellular dynamics.
identify putative driver genes and regimes of regulatory changes.
infer a latent time to reconstruct the temporal sequence of transcriptomic events.
estimate reaction rates of transcription, splicing and degradation.
use statistical tests, e.g., to detect different kinetics regimes.
scVelo has, for instance, recently been used to study immune response in COVID-19 patients and dynamic processes in human lung regeneration. Find out more in this list of application examples.
Latest news
Aug/2021: Perspectives paper out in MSB
Feb/2021: scVelo goes multi-core
Dec/2020: Cover of Nature Biotechnology
Nov/2020: Talk at Single Cell Biology
Oct/2020: Helmholtz Best Paper Award
Oct/2020: Map cell fates with CellRank
Sep/2020: Talk at Single Cell Omics
Aug/2020: scVelo out in Nature Biotech
References
La Manno et al. (2018), RNA velocity of single cells, Nature.
Bergen et al. (2020), Generalizing RNA velocity to transient cell states through dynamical modeling, Nature Biotech.
Bergen et al. (2021), RNA velocity - current challenges and future perspectives, Molecular Systems Biology.
Support
Found a bug or would like to see a feature implemented? Feel free to submit an issue. Have a question or would like to start a new discussion? Head over to GitHub discussions. In either case, you can also always send us an email. Your help to improve scVelo is highly appreciated. For further information visit scvelo.org.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file scvelo-0.2.5.tar.gz.
File metadata
- Download URL: scvelo-0.2.5.tar.gz
- Upload date:
- Size: 27.3 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.1 CPython/3.8.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 7e32d9e34245971330d69c12f4339cebe0acebb61e59a8b1aca9b369078b5207 |
|
| MD5 | 0d98dbad1515d8aa463e0386cb2d9de0 |
|
| BLAKE2b-256 | d7ca6669ad5c6765493ea9db0fb54b766d043b09db5a146f178c106b7162d1ef |
File details
Details for the file scvelo-0.2.5-py3-none-any.whl.
File metadata
- Download URL: scvelo-0.2.5-py3-none-any.whl
- Upload date:
- Size: 220.0 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.1 CPython/3.8.13
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 083b7f6babe66ddc2a7d047dcd440dcb27b521c7121c051f7c22b8b7fd8e637d |
|
| MD5 | 0bb0ff2777d3da8d154276acd883c7d8 |
|
| BLAKE2b-256 | 61a60f2e185dc3ba433c262ab423dccd26d28ae0134728a95ae84ea4cd3a9e08 |















