sMRIPrep (Structural MRI PREProcessing) pipeline
Project description





sMRIPrep is a structural magnetic resonance imaging (sMRI) data preprocessing pipeline that is designed to provide an easily accessible, state-of-the-art interface that is robust to variations in scan acquisition protocols and that requires minimal user input, while providing easily interpretable and comprehensive error and output reporting. It performs basic processing steps (subject-wise averaging, B1 field correction, spatial normalization, segmentation, skullstripping etc.) providing outputs that can be easily connected to subsequent tools such as fMRIPrep or dMRIPrep.
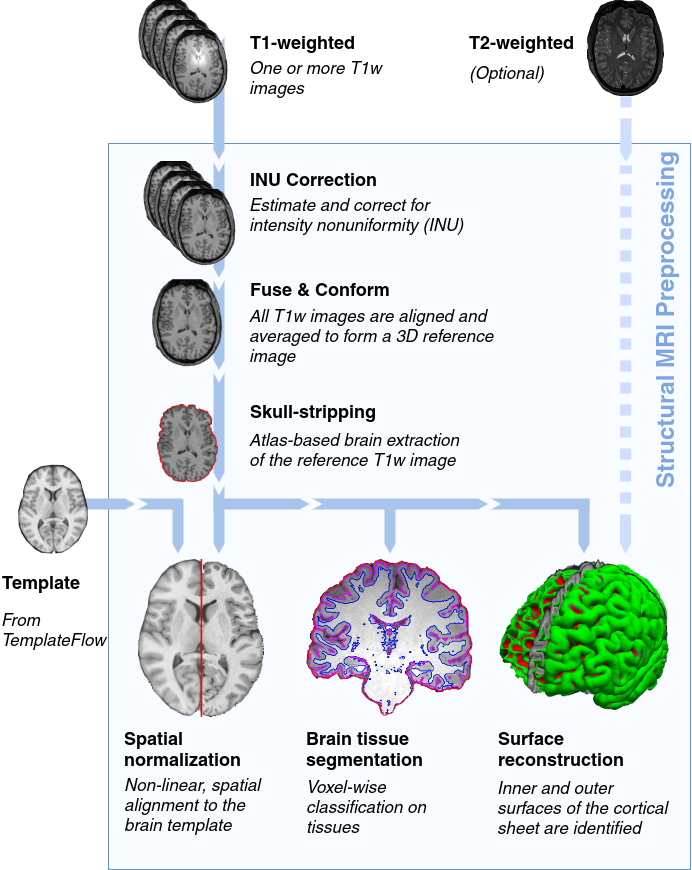
The workflow is based on Nipype and encompases a combination of tools from well-known software packages, including FSL, ANTs, FreeSurfer, and AFNI.
More information and documentation can be found at https://www.nipreps.org/smriprep/. Support is provided on neurostars.org.
Principles
sMRIPrep is built around three principles:
Robustness - The pipeline adapts the preprocessing steps depending on the input dataset and should provide results as good as possible independently of scanner make, scanning parameters or presence of additional correction scans (such as fieldmaps).
Ease of use - Thanks to dependence on the BIDS standard, manual parameter input is reduced to a minimum, allowing the pipeline to run in an automatic fashion.
“Glass box” philosophy - Automation should not mean that one should not visually inspect the results or understand the methods. Thus, sMRIPrep provides visual reports for each subject, detailing the accuracy of the most important processing steps. This, combined with the documentation, can help researchers to understand the process and decide which subjects should be kept for the group level analysis.
Acknowledgements
Please acknowledge this work by mentioning explicitly the name of this software (sMRIPrep) and the version, along with a link to the GitHub repository or the Zenodo reference (doi:10.5281/zenodo.2650521).
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file smriprep-0.10.0.tar.gz.
File metadata
- Download URL: smriprep-0.10.0.tar.gz
- Upload date:
- Size: 77.9 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.1 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 2d2058756001431dd91f050683418e6939b28ac1e52b3cc27caf85a708dd900c |
|
| MD5 | 17ac65ec34b398f12be0addec4c5c0b3 |
|
| BLAKE2b-256 | 18e4dee9adc41457cba7ec155c569eabebbadca1ce6cf6006b7577fa126b1781 |
File details
Details for the file smriprep-0.10.0-py3-none-any.whl.
File metadata
- Download URL: smriprep-0.10.0-py3-none-any.whl
- Upload date:
- Size: 5.3 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.1 CPython/3.8.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 20bff7608a4c0bcb24709d1272a627d885644f75b3ab3225426a280f917bb348 |
|
| MD5 | 4c7a0b3de8545074f07f8c17fae41218 |
|
| BLAKE2b-256 | 7c310fa1599bd19cf3d95b3f628d288be252ec710b6e6e20ab709df9f741149f |