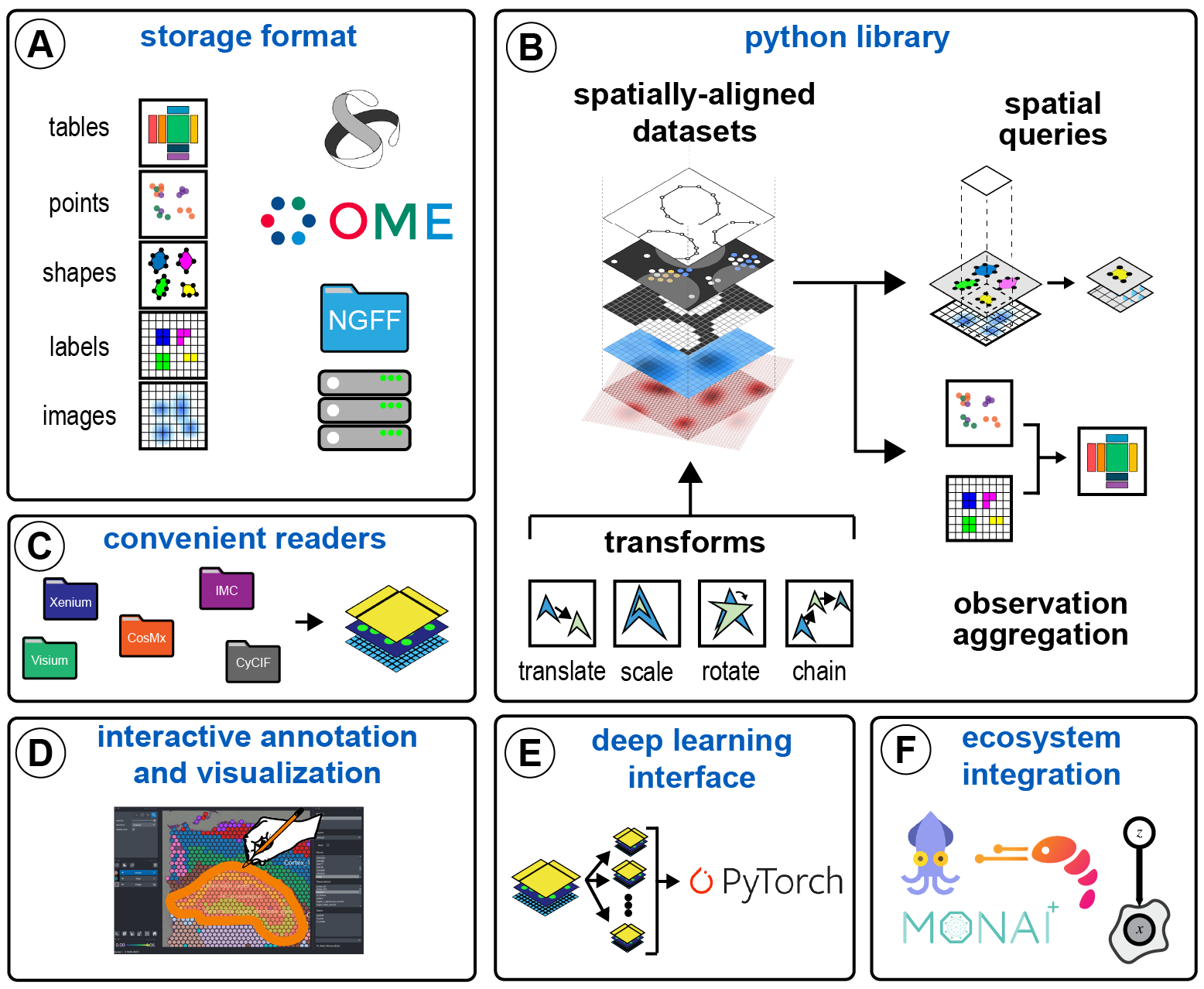
Spatial data format.
Project description
SpatialData: an open and universal framework for processing spatial omics data.
SpatialData is a data framework that comprises a FAIR storage format and a collection of python libraries for performant access, alignment, and processing of uni- and multi-modal spatial omics datasets. This repository contains the core spatialdata library. See the links below to learn more about other packages in the SpatialData ecosystem.
- spatialdata-io: load data from common spatial omics technologies into spatialdata.
- spatialdata-plot: Static plotting library for spatialdata.
- napari-spatialdata: napari plugin for interactive exploration and annotation of spatial data.
The spatialdata project uses a consensus based governance model and is fiscally sponsored by NumFOCUS. Consider making a tax-deductible donation to help the project pay for developer time, professional services, travel, workshops, and a variety of other needs.
The spatialdata project also received support by the Chan Zuckerberg Initiative.
- The library is currently under review. We expect there to be changes as the community provides feedback. We have an announcement channel for communicating these changes, please see the contact section below.
- The SpatialData storage format is built on top of the OME-NGFF specification.
Getting started
Please refer to the documentation. In particular:
Another useful resource to get started is the source code of the spatialdata-io package, which shows example of how to read data from common technologies.
Installation
Check out the docs for more complete installation instructions. To get started with the "batteries included" installation, you can install via pip:
pip install "spatialdata[extra]"
Note: if you are using a Mac with an M1/M2 chip, please follow the installation instructions.
Limitations
- Code only manually tested for Windows machines. Currently the framework is being developed using Linux, macOS and Windows machines, but it is automatically tested only for Linux and macOS machines.
Contact
To get involved in the discussion, or if you need help to get started, you are welcome to use the following options.
- Chat via
scverseZulip (public or 1 to 1). - Forum post in the scverse discourse forum.
- Bug report/feature request via the GitHub issue tracker.
- Zoom call as part of the SpatialData Community Meetings, held every 2 weeks on Thursday, schedule here.
Finally, especially relevant for for developers that are building a library upon spatialdata, please follow this channel for:
- Announcements on new features and important changes Zulip.
Citation
Marconato, L., Palla, G., Yamauchi, K.A. et al. SpatialData: an open and universal data framework for spatial omics. Nat Methods (2024). https://doi.org/10.1038/s41592-024-02212-x
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file spatialdata-0.2.0rc1.tar.gz.
File metadata
- Download URL: spatialdata-0.2.0rc1.tar.gz
- Upload date:
- Size: 200.4 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.12.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 62602a4f5b127ffd9252bd3230c1f1911d5f3d78010f6a0938501960da5940e7 |
|
| MD5 | cdd8e2a68bb12d6476e14d62ee911c3c |
|
| BLAKE2b-256 | 1ecab4cd79974ca911524398e557256ceba1dd6dd7d036b08154da4f2948b4a2 |
File details
Details for the file spatialdata-0.2.0rc1-py3-none-any.whl.
File metadata
- Download URL: spatialdata-0.2.0rc1-py3-none-any.whl
- Upload date:
- Size: 159.2 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.12.4
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | ba04235990c4b253509e42ac1cfb2c55122aa09428d608255e607d6c5a4ea9c7 |
|
| MD5 | cde437113eee3c9f6643371224517527 |
|
| BLAKE2b-256 | 37300e2a587438e070c4313d1c3d0b2e567f896508795a953d6f452110528fcf |