Tools for producing sweights using classic methods or custom orthogonal weight functions (COWs) and for corrected covariance matrices for weighted data fits.
Project description
sweights
pip install sweights
We provide a few tools for projecting component weights ("sweights") for a control variable(s) using a discriminating variable(s). For details please see Dembinski, Kenzie, Langenbruch, Schmelling - arXiv:2112.XXXXX.
We also provide tools for correcting the covariance matrix of fits to weighted data. For details please see Dembinski, Kenzie, Langenbruch, Schmelling - arXiv:2112.XXXXX (Sec IV) and Langenbruch - arXiv:1911.01303.
Documentation
A variety of options for extraction of signal-like weights is provided:
-
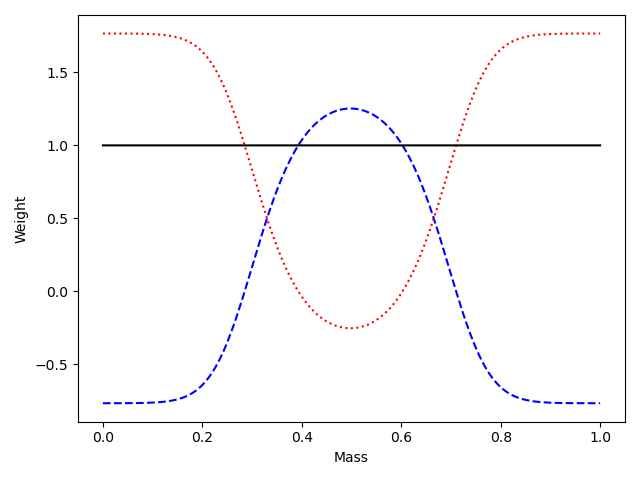
classic "sWeights" via the
sweightclass produces pure weight functions and provides the Wkl and Akl matrices- Using the "summation" method from the original sPlot paper Pivk, Le Diberder - arXiv:physics/0402083 referred to as Variant B in arXiv:2112.XXXXX
- Using the "integration" method rederived originally by Schmelling and referred to as Variant A in arXiv:2112.XXXXX
- Using the "refit" method, i.e. taking the covariance matrix of a yield only fit, referred to as Variant Ci in arXiv:2112.XXXXX
- Using the "subhess" method, i.e. taking the sub-covariance matrix for the yields, referred to as Variant Cii in arXiv:2112.XXXXX
- Using the implementation in ROOT's TSPlot (this we believe should be equivalent to Variant B but is more susceptible to numerical differences)
- Using the implementation in RooStat's SPlot (we found this identical to Variant B ("summation") above in all the cases we tried)
-
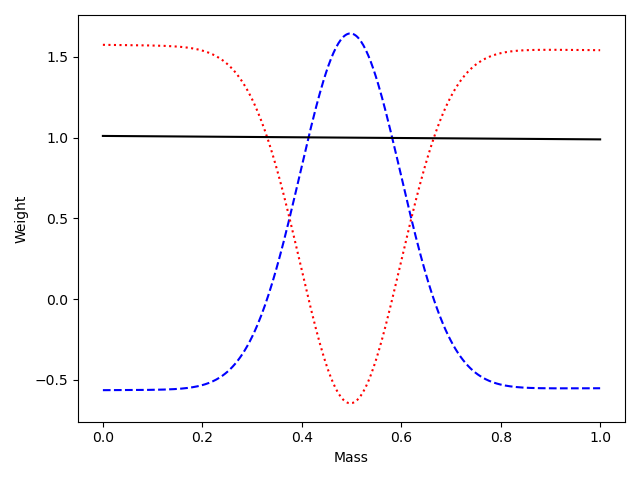
Custom Orthogonal Weight functions (COWs) via the
cowclass produces pure weight functions and provides Wkl and Akl matrices. It expects- gs - the signal function for the discrimant variable
- gb - the background function(s) for the discriminant variable (can pass orders of polynomials here if desired)
- Im - the variance function for the discriminant variance (can also pass 1 and it will be set of uniform)
- obs - one can instead or additionally pass the observed distribution in the discriminant variable which will be used for the variance function instead. In this case you must pass a two element tuple giving the bin entries and bin edges for the observed dataset (the same as what
np.histogram(data)would return)
Corrections to the covariance matrix can be implemented using
cov_correctwhich computes the full asymptotic correction using Eq. 55 in arXiv:2112.XXXXXapprox_cov_correctis more straightfoward to compute but only computes the first term in Eq. 55 of arXiv:2112.XXXXX which will be slightly conservative
A test of variable independence based on the Kendall tau coefficient is also provided in kendall_tau
Examples
An example script demonstrating a typical use case is provided in
python tests/examples.py
This does the following:
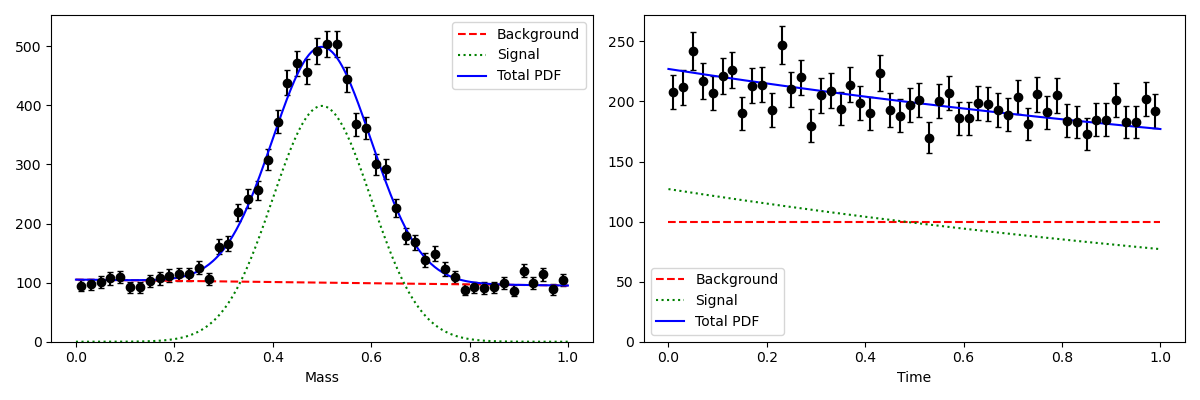
- Generate toy data in two independent dimensions m and t
-
Fit the toy data in the discriminanting variable to get an estimate of the discriminating variable pdfs
-
Compute sWeights using the "summation" method (implemented by the
sweightclass provided)
- Compute sWeights using the COW method with a variance function of unity, I(m)=1, (implemented by the
cowclass provided)
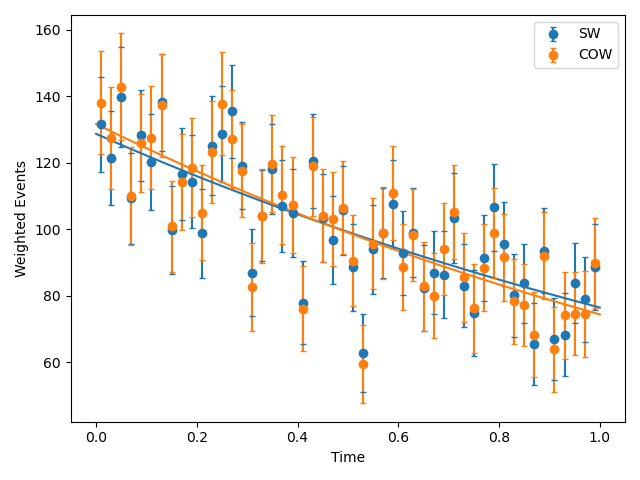
- Fit the weighted distributions and correct the covariance using the
cov_correctfunction provided
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file sweights-0.0.4.tar.gz.
File metadata
- Download URL: sweights-0.0.4.tar.gz
- Upload date:
- Size: 16.0 kB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.6.0 importlib_metadata/4.8.2 pkginfo/1.8.1 requests/2.25.0 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.8.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | d18232f6a3861b76af84e4dd01e34b33a0df145e88369629b7e3ae9bb5403f02 |
|
| MD5 | 86a965ec2cd961686435b5f4424a98e7 |
|
| BLAKE2b-256 | ee87a921f7986ad4d32b9bde3cbc9ec8f14d97f930d744239411f7bde1814eb6 |
File details
Details for the file sweights-0.0.4-py3-none-any.whl.
File metadata
- Download URL: sweights-0.0.4-py3-none-any.whl
- Upload date:
- Size: 15.7 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.6.0 importlib_metadata/4.8.2 pkginfo/1.8.1 requests/2.25.0 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.8.6
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 18078ebd2f5b383c11dd27145f7c793eb13befe851a733f7721016ac0788ae46 |
|
| MD5 | 1e24de7800a62162a82cab47b97a4dc2 |
|
| BLAKE2b-256 | 1c0255b7bfea86c17486ae52c2163511c54a56bb7f3fb5699802ff44221b6fbc |