A non-interacting equilibrium 2D quantum transport simulation framework
Project description
Transport In a Non-Interacting Equilibrium simulation framework (TINIE)
Overview
We present to you a code that calculates conductance and electric current running through 2D cavities, quantum dots or potential wells with arbitrarily placed reservoirs in a perpendicular and constant magnetic field. The code can be used in a wide range of calculations involving 2D electron transport. The main difference between this code and its competitors is the fact that TINIE does not need to be provided with free parameters. The code is parallelized with mpi4py, allowing for computational tasks distribution across multiple processors.
Installation
The code is a Python package (written using Python 3.6). In non-parallel simulations, tinie can function without MPI-support in the HDF5-library, but parallel simulations require the HDF5 library and its Python-wrapper h5py to be built with MPI support (see Parallel HDF5 – h5py documentation for installation instructions).
tinie can be installed with pip:
$ pip install tinietinie installs a command line interface toolset, and you can test the functionality with the following commands:
$ tinie_prepare
$ tinieThorough test suite for the package has been implemented and can be launched via python3 setup.py test from the root of the git repository.
Package Functionality
This package contains tools that could be used to calculate coupling of a specific system that contains a central region (2DEG) and some leads. After the coupling is calculated one could proceed to calculate transmission coefficients and partial currents in the leads. All the calculations are performed in Hartee atomic units. After installing the package, a simple test run can be launched as follows:
$ tinie_prepare
$ tiniePackage Structure
The code is written using object-oriented programming, and its
functionality can be shortly described in the following way: first,
Lead, and Center objects are created and passed as inputs for
the Coupling object and then all of them are passed into a
SystemDump object, which calculates all the couplings and
Hamiltonians and dumps the data into an hdf5 file. SystemFetch is
then used to read the data from that hdf5 file. That data is passed into
the Calculator interface, where SelfEnergy interface calculates
the self-energies 

After that, SelfEnergy for all the leads and the eigenenergies of
the Center are passed into the GreenFunction interface that
evaluates the advanced (


in the lead, where 
Modular structure of the code allows for the implementation of your own custom type of conducting channel (“lead”), quantum dot (“central region”) and coupling via implementation of a class that inherits from Lead, Center or Coupling. The details of how exactly the classes should be implemented will be explained in the sections to follow.
Example: Usage of TINIE with ITP2D
To better demonstrate how TINIE is used, we will show it by means of an example problem. We will compute transport properties of a quantum-dot system with two leads in a magnetic field. Specifically, we shall procure the information about the central region from ITP2D, a Schrödinger equation eigensolver that interfaces with TINIE. The following workflow is typical for most transport problems solved with TINIE:
Step 0: computation of the Hamiltonian and wavefunctions of the
central region. We may obtain this information from any eigensolver of
our choosing, provided that it is TINIE-compatible. Quantum dot may be
modeled by a radial harmonic potential of form 

$ itp2d -v -n 20 -l 12 -s 100 -p "harmonic(1)" -B 1.0 -o ITP2D_FILE_PATHHere the central region occupies a 
Step 1: the transport system preparation step. In this step, the
coupling matrices for the leads are computed. Suppose we wish to compute
overlap coupling between the central region and the leads and we want to
vary the probe energy within each lead in range 

Lead 0 is confined to region
in x-direction,
in y-direction and connects to the lead from the left;
Lead 1 is confined to region
in x-direction,
in y-direction and connects to the lead from the right.
Both leads in this case have harmonic potential of strength

$ tinie_prepare -dE 1e-3 -B 1.0 -ctr "itp2d(ITP2D_FILE_PATH,(0,4))" -l 2 -ld "finharm(left,1.0,dir)" "finharm(right,1.0,dir)" -xlim "[-10.0,-4.0]" "[4.0,10.0]" -ylim "[-5.0,5.0]" "[-5.0,5.0]" -Elim "[0.0,2.0]" "[0.0,2.0]" -cpl "overlap()" "overlap()" -o TINIE_PREPARE_FILE_PATHThis produces the PREPTINIEFile that contains the information about the coupling of the transport system which can be reused for different transport calculations of the next step.
Step 2: the transport calculation step. This is where the real fun
begins, the steps before are in a sense just a preparation. To compute
various transport properties of the system, such as transmission,
conductance and current, we fix temperature of the system






$ tinie -i TINIE_PREPARE_FILE_PATH -dw 1e-2 -eta 1e-1 -T 1.0 -mu 1.0 -V 0.5 1.5 -o TINIE_FILE_PATHThis produces the TINIEFile that contains all the above mentioned transport quantities and more, with detailed description of its contents outlined in the sections below.
In addition to the transport properties, we can compute local and standard density of states (LDOS/DOS) of the system via the tinie_dos script. To that end, in addition to the parameters specified above, user would want to specify the energies at which LDOS should be evaluated, as well as the location of the file with the central region wavefunctions. We then use the script as follows:
$ tinie_dos -i TINIE_PREPARE_FILE_PATH -psi ITP2D_FILE_PATH --wf-range 0 4 -w 1.0 2.0 3.0 -dw 1e-2 -eta 1e-1 -T 1.0 -mu 1.0 -V 0.5 1.5 -o TINIE_DOS_FILE_PATHHere, we evaluated LDOS at probe energies

Step 3: visualizing the results. To that end, one can use the
tinie_draw script. Suppose we want to plot transmission,
conductance, total current and DOS of the system in the energy range


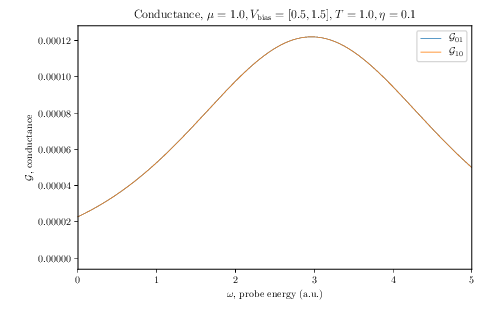
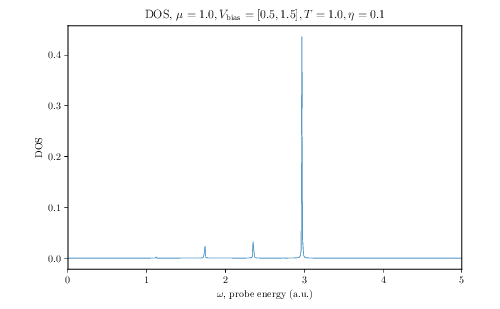
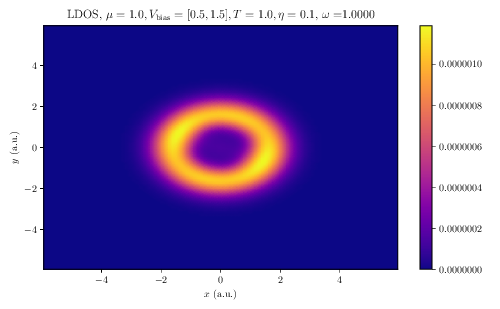
$ tinie_draw -i TINIE_FILE_PATH -idos TINIE_DOS_FILE_PATH -E 0.0 5.0 --ldos-E 1.0 --transmission --conductance --total-currents --dos --ldos -o FIGURE_PATHThis will produce beautiful LaTeX-rendered plots of the aforementioned quantities. Below we show the example plots of conductance, DOS and LDOS produced by the script:
Conductance |
DOS |
LDOS |
|---|---|---|
Quantum transport calculations in two-dimensional systems have never been this easy!
Currently Implemented System Classes
As of now, the following system classes are implemented:
Center objects, located in transport_calculator/systems/central_region
Itp2dCenter: itp2d-compatible interface.
CustomCenter: container for a custom predefined central region Hamiltonian
.
Lead objects, located in transport_calculator/systems/leads. Note that wavefunction normalization has been omitted for the sake of compactness of the expression. Wavefunctions in the code are all normalized.
FiniteHarmonicLead: lead described by a wavefunction
, where
is the the Hermite polynomial of order l,
,
,
with
being the frequency of quantum harmonic oscillator and
being magnetic field strength. The formula is provided in Hartee atomic units. x and y coordinate wavefunctions are interchangeable depending on the lead alignment.
BoxLead: particle in a box lead describe by wavefunction
where
and
are the length and width of the box correspondingly and
.
CustomLead: container for a custom predefined lead region Hamiltonian
Coupling objects, located in transport_calculator/systems/couplings
OverlapCoupling: strong coupling of the type
, where
is the ith eigenfunction of the lead and
s the jth eigenfunction of the central region and
is the overlap region of the lead and the quantum-dot.
TightBindingCoupling: weak coupling between non-overlapping lead and central regions of the type
where
,
is the lead region to be coupled and
is the central region to be coupled.
OneLayerCoupling: weak coupling between the boundaries of a non-overlapping lead and central regions of the type
.
CustomCoupling: container for custom predefined coupling matrix
. Compatible only with CustomCenter and CustomLead objects.
The implementational details of these elements can be checked in the source code, which is rich with insightful and helpful comments.
Adding Your Own Custom System Classes
As it has been mentioned before, the code has been designed in such a way as to allow as much freedom in expansion as possible. In particular, you can introduce additional types of central regions, lead regions and coupling methods. All you have to do is to create your own class file in the corresponding folder in tinie/systems and make sure that the class you are creating inherits from one of the basic abstract classes (Center, Lead or Coupling). Below you can find a list of functions you would have to implement (correctly) in order for your custom class to be fully integrated into the transport scheme:
Center region:
__init__(*attrs): initializer
get_type_specific_parameters(): retrieves child-specific extra parameters
get_energies(): retrieves central region Hamiltonian
get_potential(): retrieves potential energy values in the central region
get_state(n): retrieves nth wavefunction
get_states(): retrieves all wavefunctions on the grid
get_number_of_states(): retrieves the number of states in the central region
get_sliced_state(n, width, side): retrieves nth wavefunction on a grid slice
get_sliced_states(width, side): retrieves all wavefunctions on a grid slice
get_boundary_state(n, side): retrieves nth wavefunction evaluated on some boundary
get_coordinate_ranges(): retrieves x and y coordinate ranges
get_coordinates(): retrieves the coordinate meshes
get_slice_coordinates(width, side): retrieves the sliced coordinate meshes
get_boundary_coordinates(side): retrieves the boundary coordinate range
Lead region:
__init__(*attrs): initializer
set_magnetic_field_strength(B): sets magnetic field strength
set_energy_spacing(delta_E): sets lead energy spacing
get_type_specific_parameters(): retrieves child-specific extra parameters
get_energies(): retrieves lead region Hamiltonian
get_state_point(x, y, n): evaluates the nth state wavefunction at a single point
get_state(x_points, y_points, n, mode): retrieves the nth state wavefunction on a custom/discretized grid
get_number_of_states(): retrieves the number of states in the lead region
get_boundary_state(n, num_boundary_points): retrieves nth wavefunction evaluated on the lead boundary
get_boundary(num_boundary_points): retrieves the boundary grid with specified discretization
Coupling region:
__init__(Center_object, Lead_object, *attrs): initializer, sets the center and lead objects ready for the coupling matrix calculations
get_coupling_matrix_element(i, j): retrieves coupling matrix element
, that is, the coupling between ith lead state and jth central state
get_coupling_matrix(): retrieves the coupling matrix
Details about the input/output parameter types can be found in the source code. Upon implementing all of these functions correctly for the corresponding custom object and extending the parser interface accordingly, the code extension will be fully consistent with the original code!
Scripts Included in the Package
tinie includes a few scripts that should ease the usage of the software:
tinie_prepare
This script prepares the coupling system and saves it in a tinie_prepare hdf5 file, which contains the following attributes and datasets:
Attribute |
Description |
|---|---|
type |
File type, must be “PREPTINIEFile” |
center/type |
Type of the central region |
center/num_states |
Number of states in the central region |
center/parameters |
Type-dependent parameters of the central region |
leads/num_leads |
Number of leads |
leads/lead_n/type |
Type of the lead n |
leads/lead_n/num_states |
Number of states in lead n |
leads/lead_n/energy_spacing |
Energy spacing in lead n |
leads/lead_n/parameters |
Type-dependent parameters of lead n |
couplings/num_couplings |
Number of couplings |
couplings/coupling_n/type |
Type of coupling between lead n and the central region |
Dataset |
Description |
|---|---|
center/hamiltonian |
Hamiltonian of the central region |
center/potential |
Potential energy values in the central region |
leads/lead_n/hamiltonian |
Hamiltonian of the lead region n |
leads/lead_n/x_axis_limits |
x-axis limits of lead n |
leads/lead_n/y_axis_limits |
y-axis limits of lead n |
leads/lead_n/energy_limits |
Energy limits of lead n |
couplings/coupling_n/coupling_matrix |
Coupling matrix between lead n and the central region |
Some of these datasets are stored in chunked/compressed format for more data-intensive simulations. All the simulation parameters are adjusted via a parser user interface, which takes the following arguments (type tinie_prepare --help if you ever feel lost!):
Argument |
Description |
|---|---|
-dE, --delta-E |
Lead energy spacing |
-B |
Magnetic field strength |
-xlim, --x-axis-limits |
x-axis limits of each lead, typed in form [x_min_0, x_max_0] [x_min_1, x_max_1] ... |
-ylim` --y-axis-limits |
y-axis limits of each lead, typed in form [y_min_0, y_max_0] [y_min_1, y_max_1] ... |
-Elim --energy-limits |
Energy limits of each lead, typed in form [E_min_0, E_max_0] [E_min_1, E_max_1] ... |
-ctr --center-type |
Central region type, typed in as "ctr_type(*ctr_params)" |
-l --lead-number |
Number of leads |
-ld --lead-types |
Lead region types, typed in form "ld0_type(*ld0_params)" "ld1_type(*ld1_params)" ... |
-cpl` ``--coupling-types |
Coupling region types, typed in form "cpl0_type(*cpl0_params)" "cpl1_type(*cpl1_params)" ... |
-o, --output-file |
Path, where preptinie file is saved |
tinie
This script reads the preptinie hdf5 file, performs the transport calculation and saves the results in a tinie hdf5 with the following attributes and datasets:
Attribute |
Description |
|---|---|
type |
File type, must be “TINIEfile” |
evaluated_chemical_potential |
Chemical potential |
evaluated_bias_voltage |
Bias voltage in the leads of the system |
evaluated_temperature |
Temperature of the system |
omega_spacing |
Probe energy spacing |
lead_energy_spacing |
Lead energy spacing |
eta |
Small number |
number_of_couplings |
Number of couplings in the system |
Dataset |
Description |
|---|---|
partial_currents |
Matrix of partial currents between each lead |
total_currents |
Total currents in each lead |
omega_dependent_partial_currents |
Energy profile of the partial current matrix |
omega_ldos_dependent_total_currents |
Energy profile of the total currents |
transmission |
Transmission matrix as a function of the probe energy |
transmission_error |
Imaginary component of transmission |
conductance |
System conductance matrix |
Some of these datasets are stored in chunked/compressed format for more data-intensive simulations. All the transport calculation parameters are adjusted via a parser user interface, which takes the following arguments (type tinie --help if you ever feel lost!):
Argument |
Description |
|---|---|
-dw`, ``--delta-omega |
Probe energy spacing |
-eta |
Small imaginary constant used in calculating Green’s function |
--mu, --chem-pot |
Chemical potential at which the system is evaluated |
-V, --lead-bias |
Lead biases, at which the system is evaluated, typed in form V_0 V_1 ... |
-T, --temperature |
Temperature at which the system is evaluated |
-i, --input-file |
Path to the tinie_prepare’s output file |
-o, --output-file |
Path for the output file |
--wide-band, --no-wide-band |
Boolean flags user can specify if they wish to use the wide band approximation methods (or not) |
-S, --self-energy |
Path from which an array of custom self energies is read |
-G, --rate-operator |
Path from which an array of custom rate operators is read |
Note that if you wish to use the wide band approximation approach you must specify either self energies or rate operators or both!
tinie_dos
This scripts reads the preptinie hdf5 file and the file containing the eigenfunctions of the central region, computes DOS/LDOS and saves the results in dostinie hdf5 file with the following attributes and datasets:
Attribute |
Description |
|---|---|
type |
File type, must be “TINIEDOSFile” |
Dataset |
Description |
|---|---|
dos |
Density of states values |
ldos |
Local density of states values |
x |
x-axis values of the system central region |
y |
y-axis values of the system central region |
omega_dos |
Energies at which DOS was evaluated |
omega_ldos |
Energies at which LDOS was evaluated |
Some of these datasets are stored in chunked/compressed format for more data-intensive simulations. All the DOS/LDOS calculation parameters are adjusted via a parser user interface, which takes the following arguments (type tinie_dos --help if you ever feel lost!):
Argument |
Description |
-w, --omega-ldos |
Probe energies at which the LDOS should be avaluated |
-dw, --delta-omega |
Probe energy spacing |
-eta |
Small imaginary constant used in the calculation of the Green’s function |
--mu, --chem-pot |
Chemical potential at which the system is evaluated |
-V, --lead-bias |
Lead biases at which the system is evaluated, typed in as V_0 V_1 ... |
-T, --temperature |
Temperature at which the system is evaluated |
-i, --input-file |
Path to the tinie_prepare’s output file |
--psi, --wf-file |
Path from which central region wavefunctions are read |
-o, --output-file |
Path where the tinie_dos` file is saved |
--dos, --no-dos |
Boolean, decides if DOS is computed |
--ldos, --no-ldos |
Boolean, decides if LDOS is computed |
tinie_draw
This script reads data from the tinie hdf file, makes pretty transmission/backsacttering/current/density of states plots and saves them. This script has a parser user interface, where you can specify the following plot arguments:
Argument |
Description |
|---|---|
-i, --input-file |
Path to the tinie-file |
-idos, --input-dos-file |
Path to the tinie_dos-file |
-o, --output-file |
Basepath for plots |
-E, --energy-rangs |
Range of energies over which to draw the figures |
--transmission, --no-transmission |
Boolean, decides if transmission is plotted |
--backscattering, --no-backscattering |
Boolean, decides if backscattering is plotted |
--partial-currents, --no-partial-currents |
Boolean, decides if partial currents are plotted |
--total-currents, --no-total-currents |
Boolean, decides if total currents are plotted |
--dos, --no-dos |
Boolean, decides if DOS is plotted |
--ldos, --no-ldos |
Boolean, decides if LDOS is plotted |
--norm-ldos, --no-norm-ldos |
Boolean, decides if the LDOS will be normalized to 1 |
--ldos-E |
Evaluate LDOS at an index corresponding to a probe energy |
--stability, --no-stability |
Boolean, decides if the numerical stability tests are plotted |
make_system_files
This script generates custom Hamiltonians or coupling matrices and saves them in a .npy file to be passed on as arguments for CustomCenter/CustomLead/CustomCoupling objects. They can also be used to generate custom self-energy/rate operators for the wide-band approximation. Run the script, follow the instructions and the rest is history.
Naturally, these scripts provide only some of the basic functionality extensions. Additional scripts/code modifications may be added based on the user’s end goals.
Project details
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file tinie-1.0.6.tar.gz.
File metadata
- Download URL: tinie-1.0.6.tar.gz
- Upload date:
- Size: 16.4 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.2.0 pkginfo/1.5.0.1 requests/2.24.0 setuptools/47.3.1 requests-toolbelt/0.9.1 tqdm/4.47.0 CPython/3.7.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | c9374487f36e6d636a0c03026a966a4f976b789252c545dbc1498c6cf41bf39b |
|
| MD5 | 1f1d4f774b3a88eada8ebc622b5d3b23 |
|
| BLAKE2b-256 | 1af3ed69f8dbbe6c56e8f608702ca49351b5f0a88437760375180544fe9cf487 |
File details
Details for the file tinie-1.0.6-py3-none-any.whl.
File metadata
- Download URL: tinie-1.0.6-py3-none-any.whl
- Upload date:
- Size: 16.5 MB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.2.0 pkginfo/1.5.0.1 requests/2.24.0 setuptools/47.3.1 requests-toolbelt/0.9.1 tqdm/4.47.0 CPython/3.7.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | e09eb668289e127f8586f3063dc412d599c1fadf00dbc5a40d69f98074fb729a |
|
| MD5 | eb97e8d58b38f4313bf6d32f914246d2 |
|
| BLAKE2b-256 | c9391d5c1d3d3200753e549ad256b6b0eefe783d7f33695ca959f458c4e4da13 |















 of the system
of the system used in the Green’s function
used in the Green’s function








