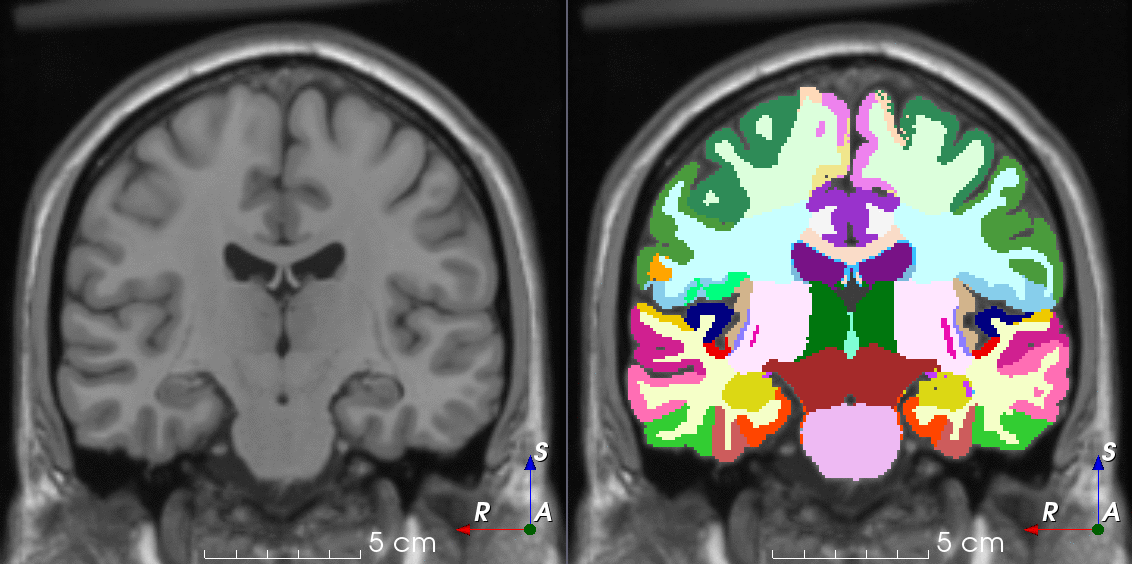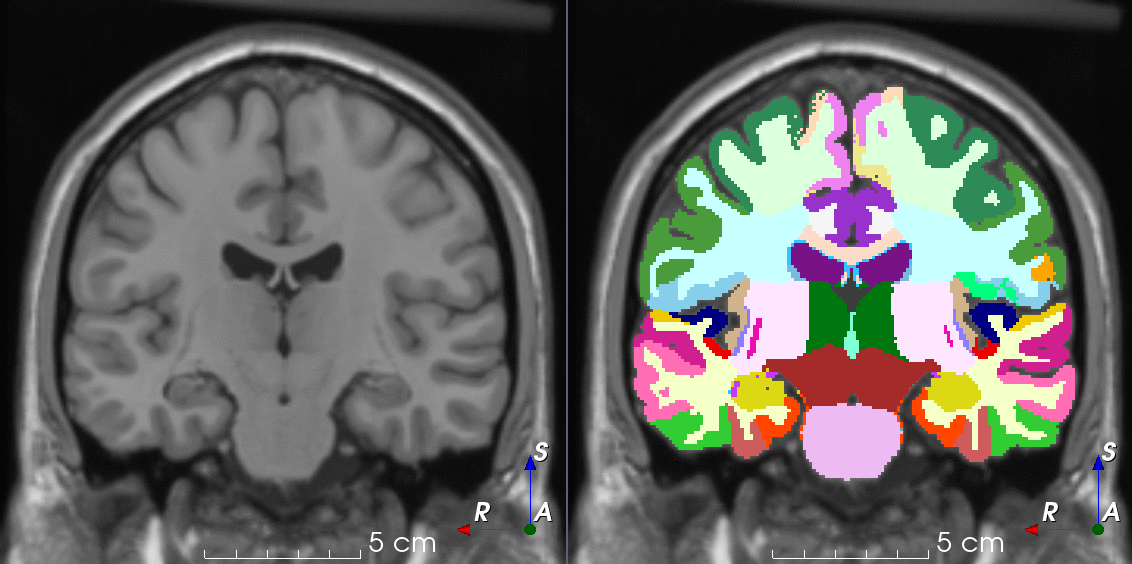
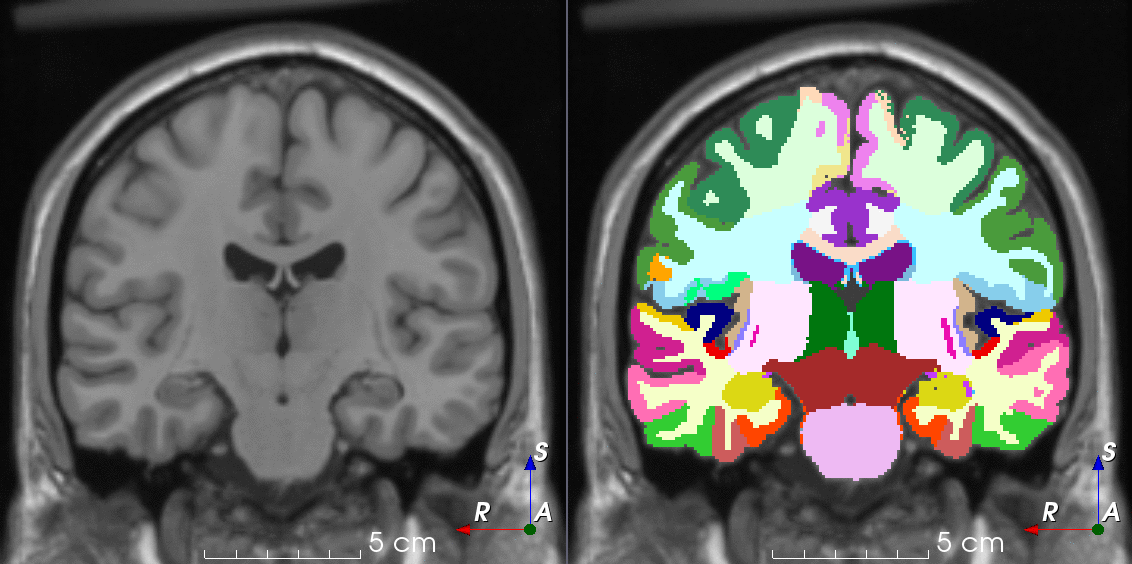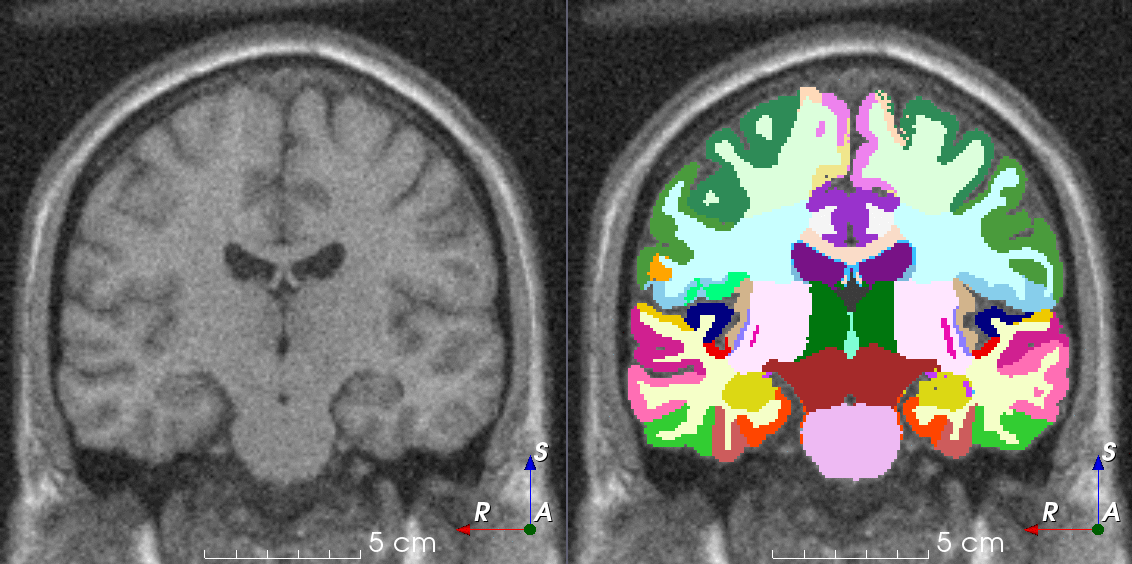
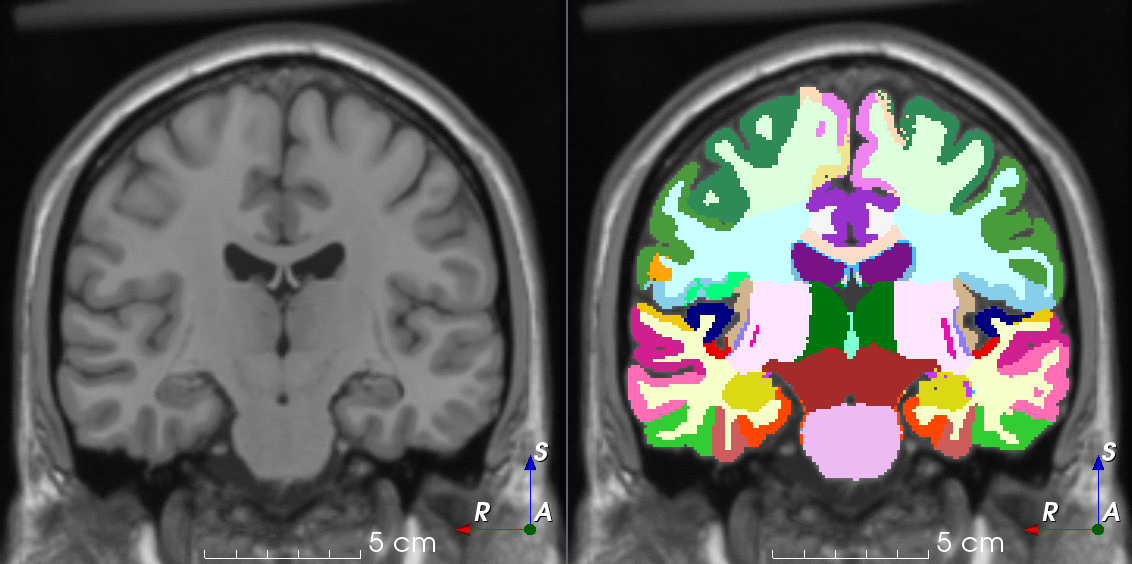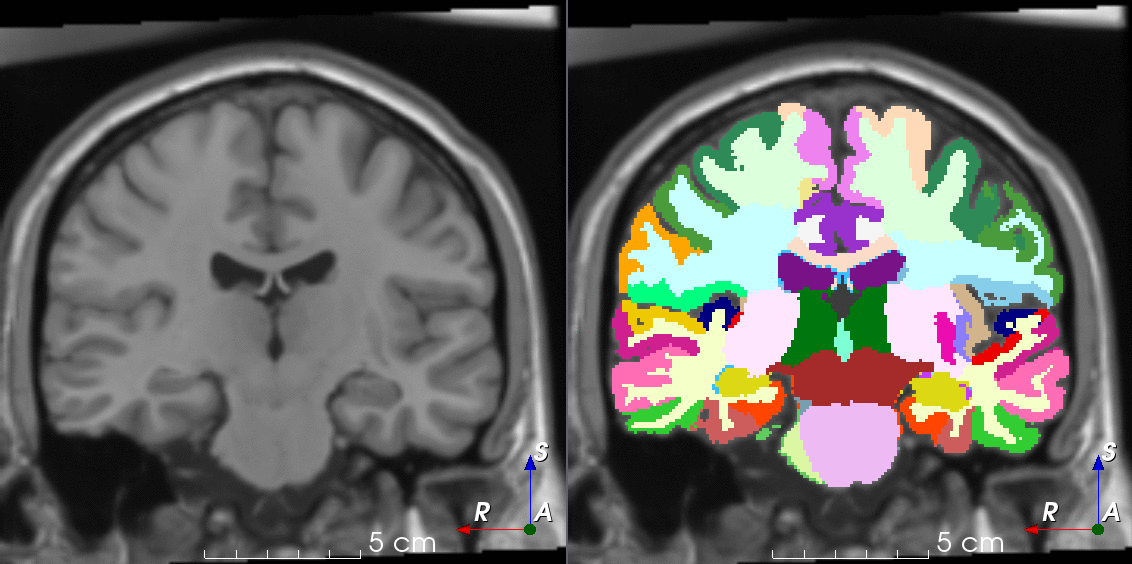
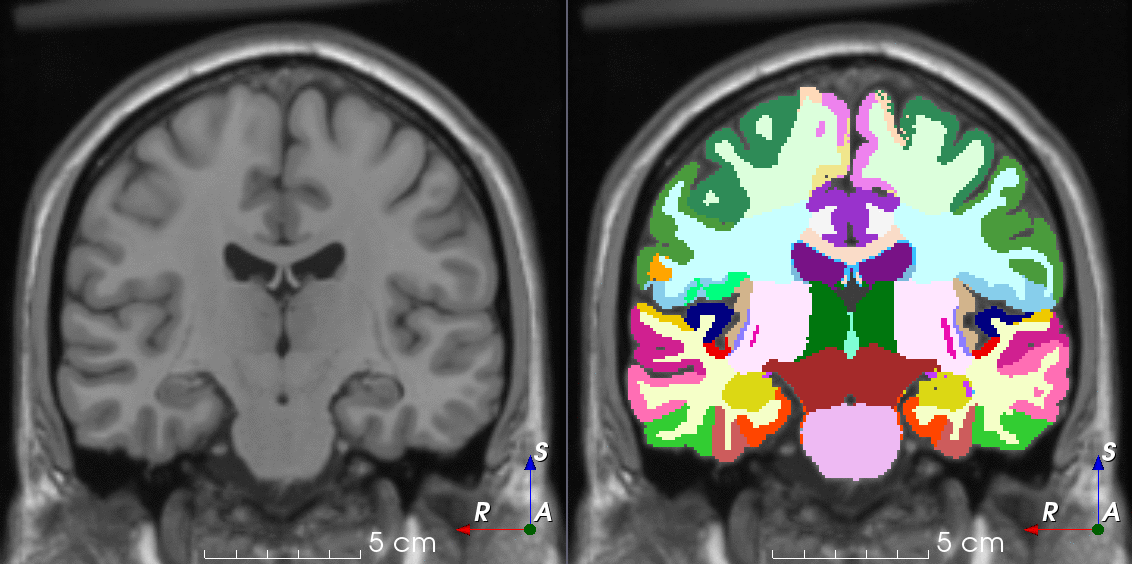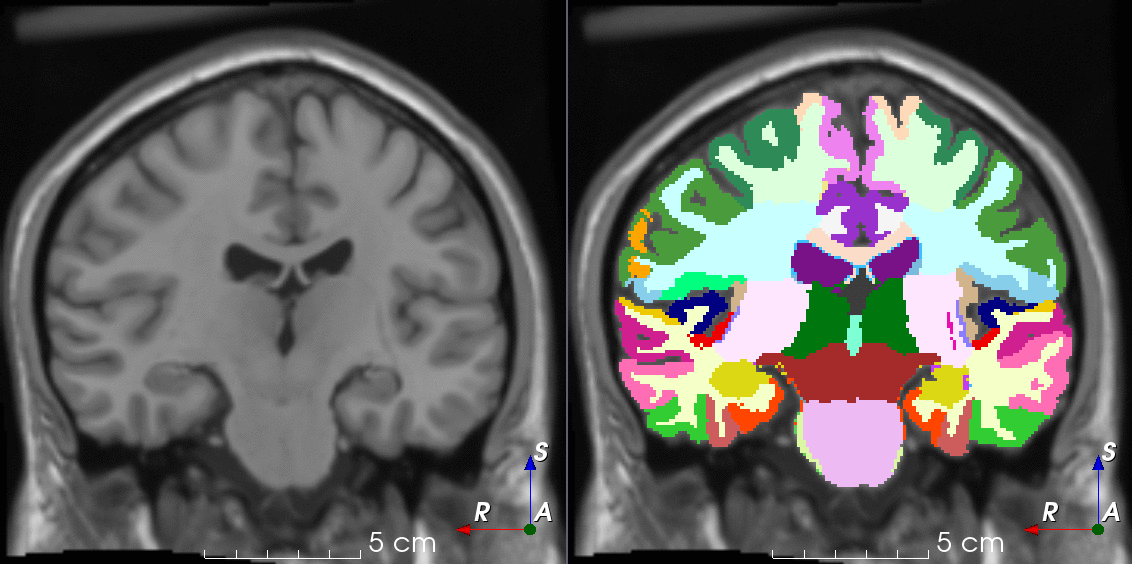
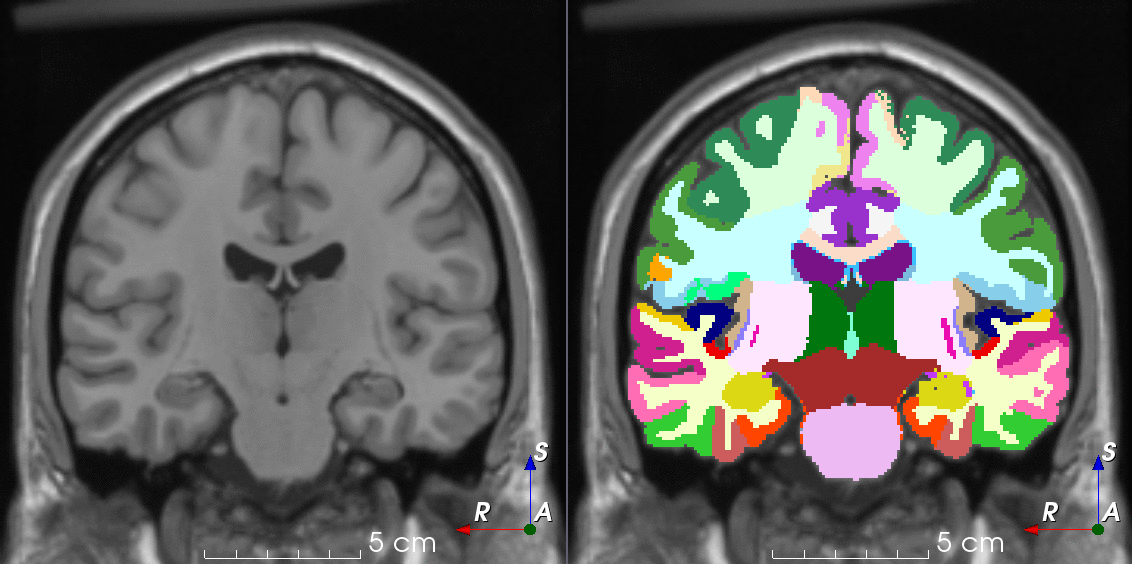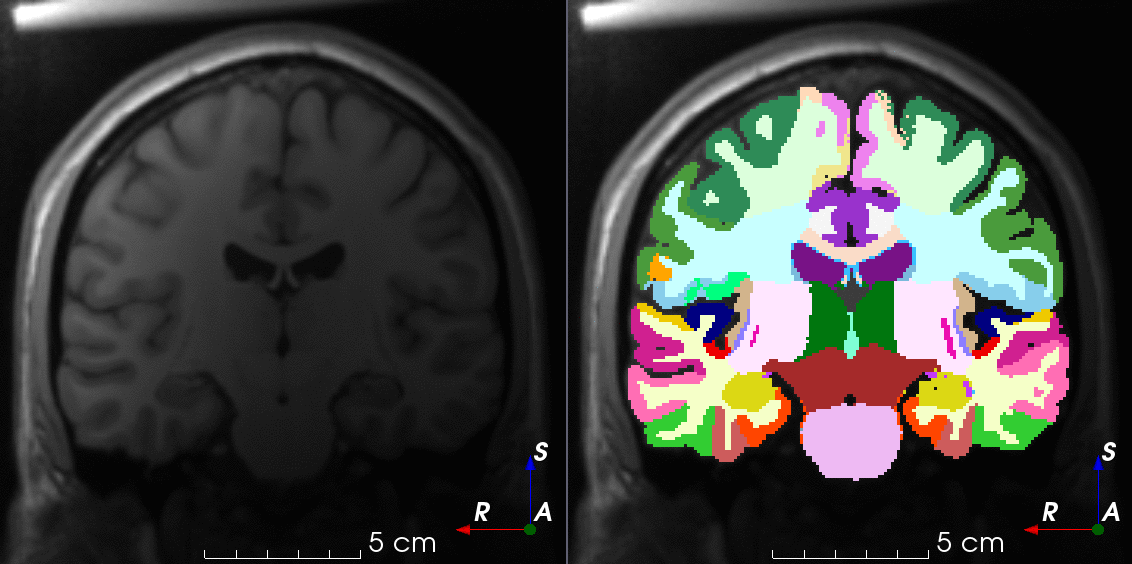
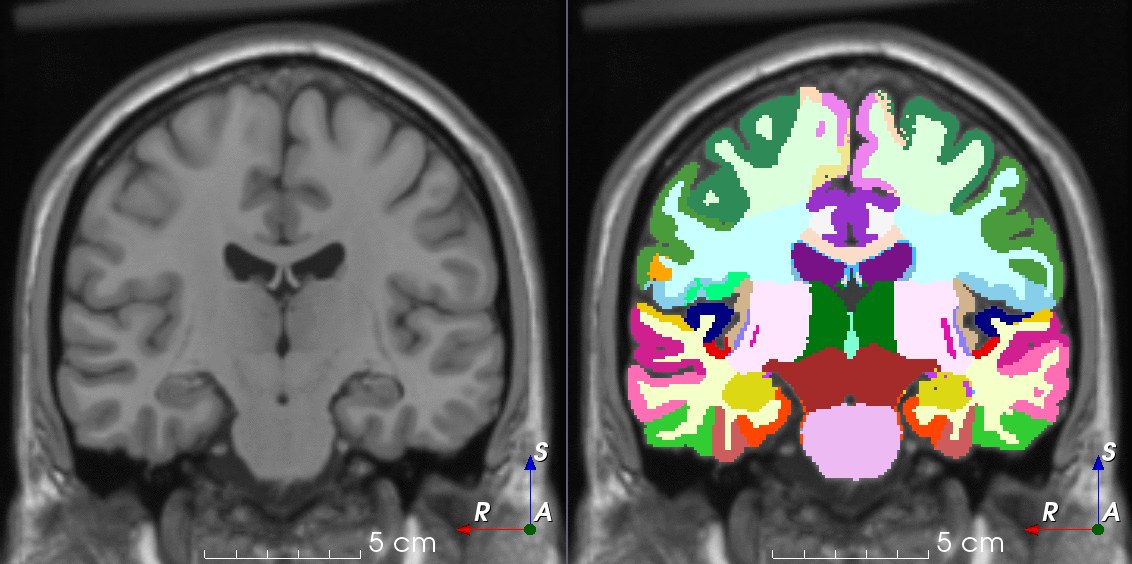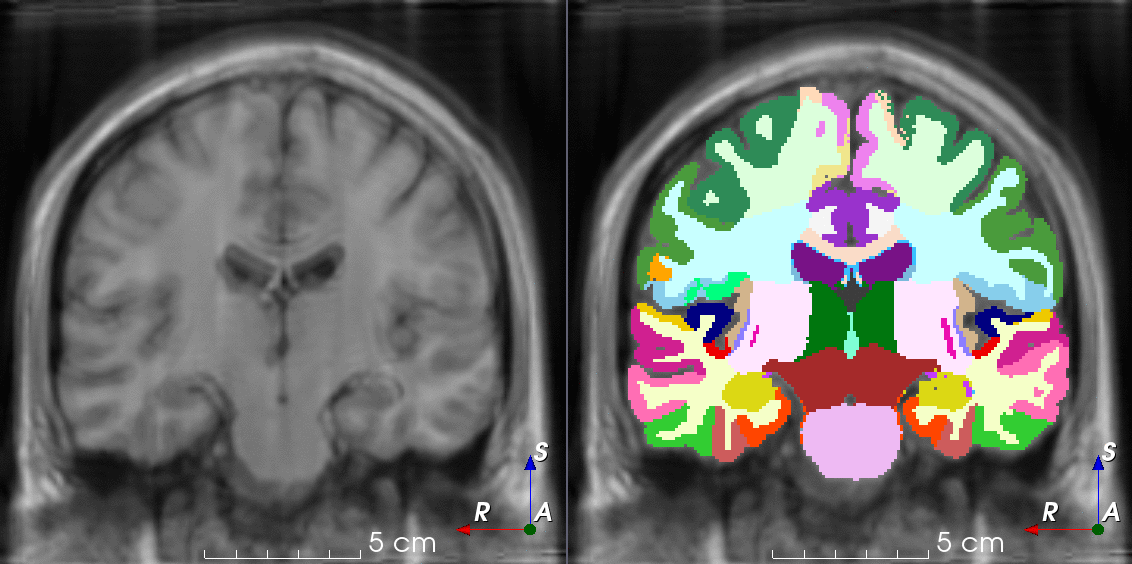
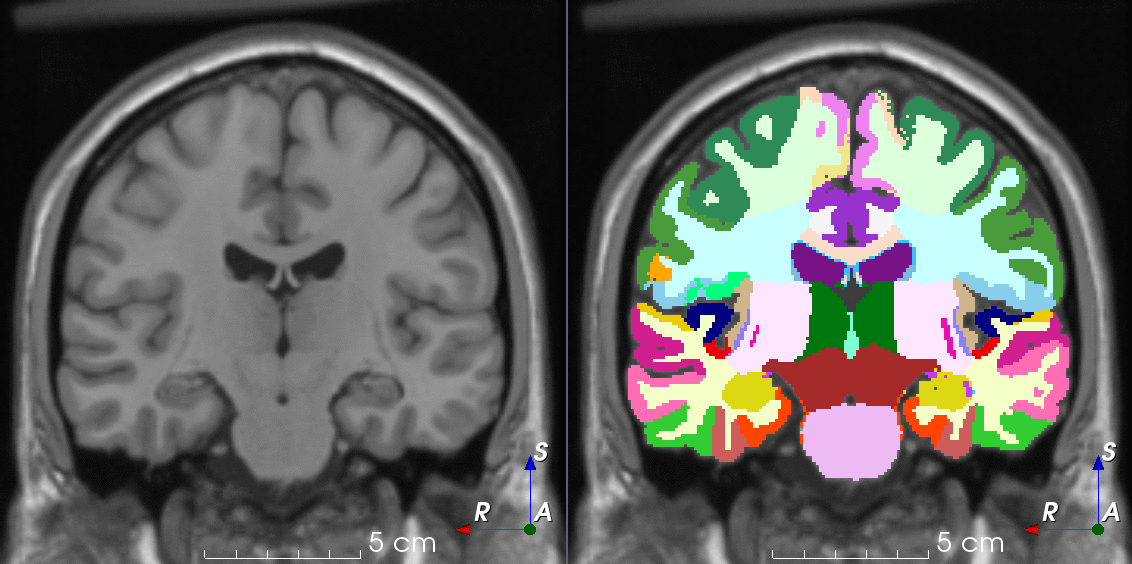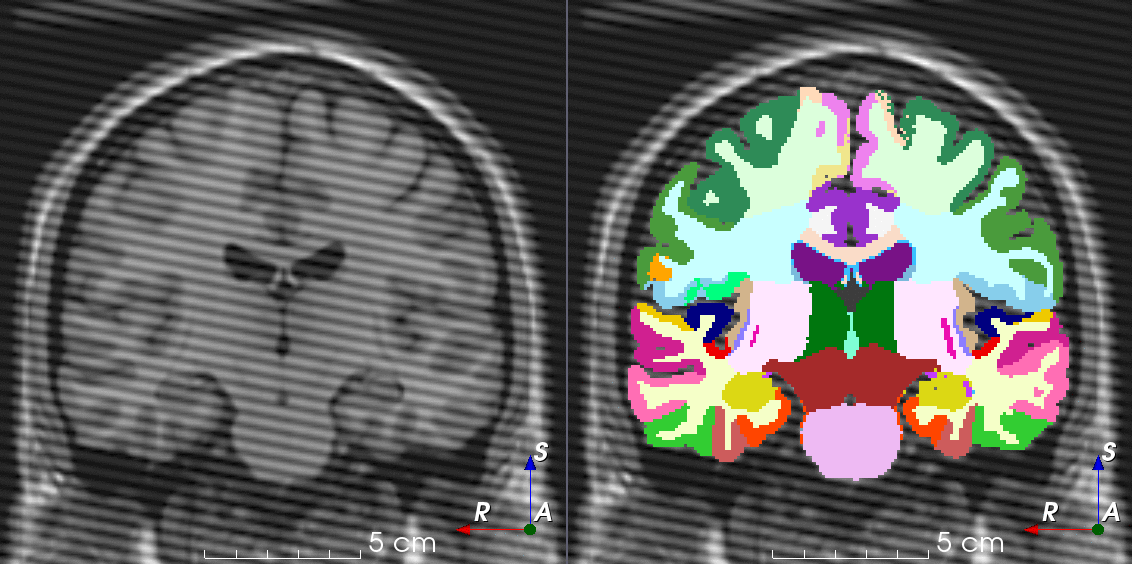
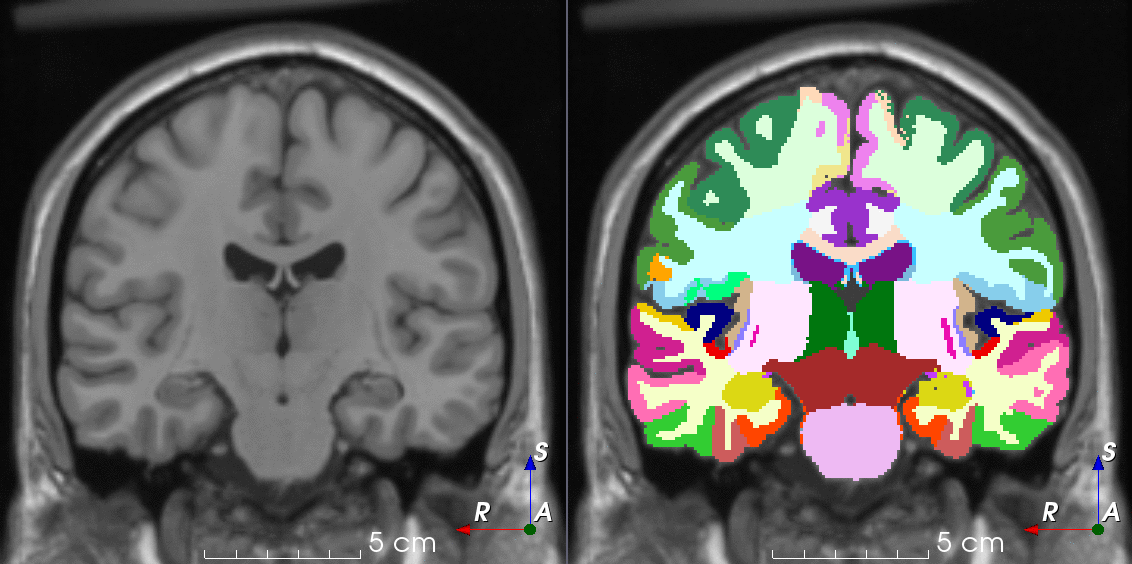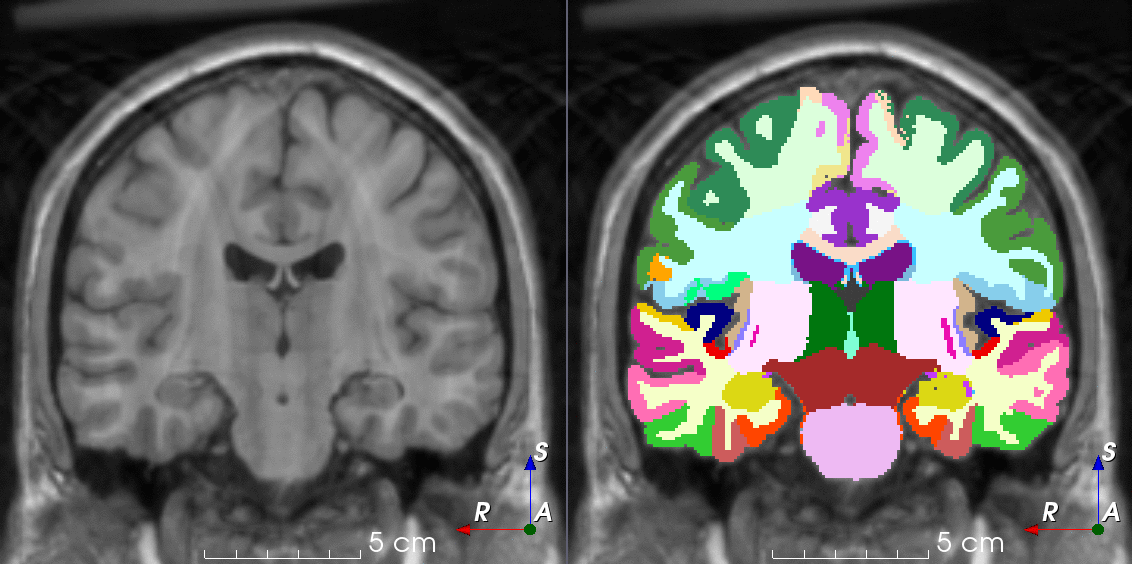
Tools for loading, augmenting and writing 3D medical images on PyTorch.
Project description
Tools like TorchIO are a symptom of the maturation of medical AI research using deep learning techniques.
Jack Clark, Policy Director at OpenAI (link).
| Package |



|
| CI |




|
| Code |



|
| Tutorials |

|
| Community |





|
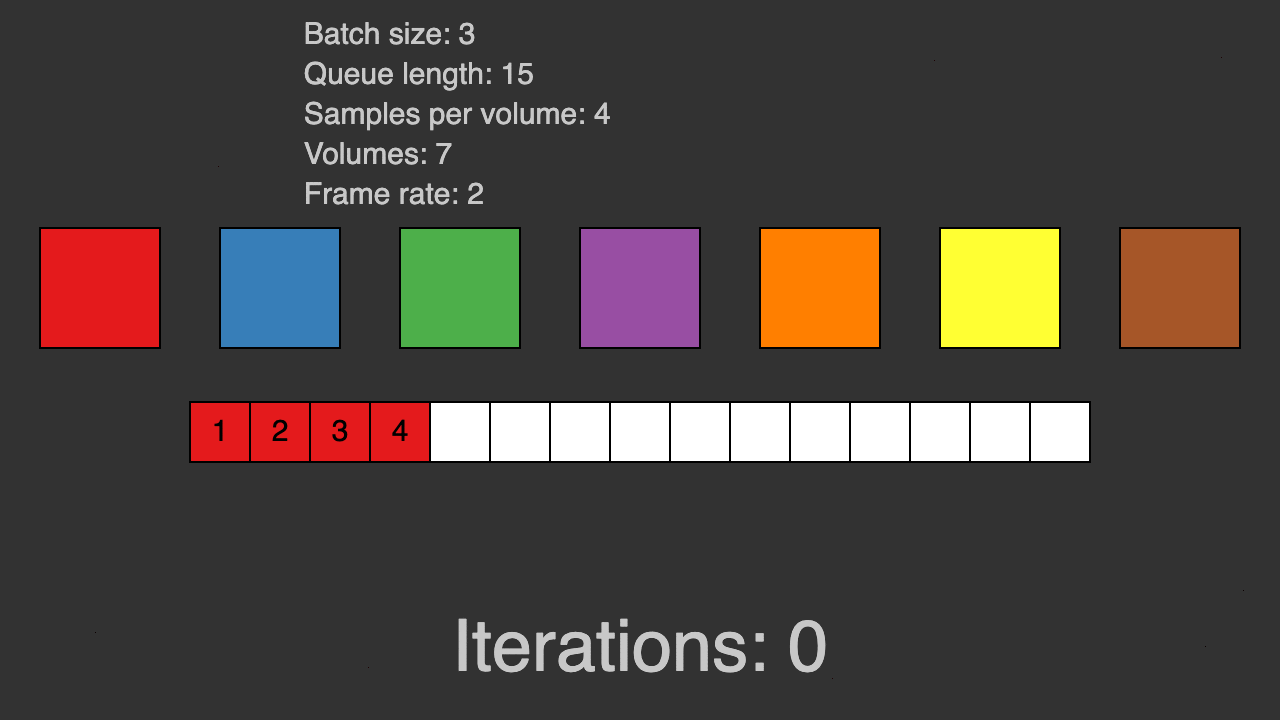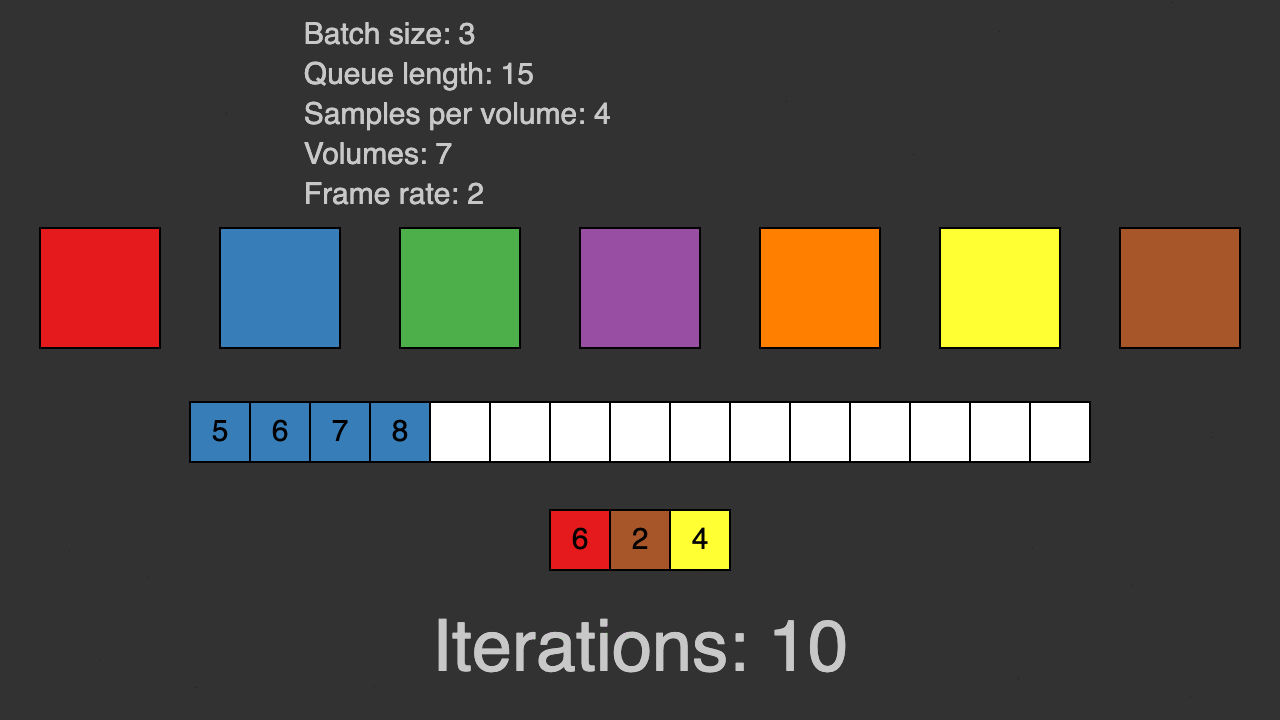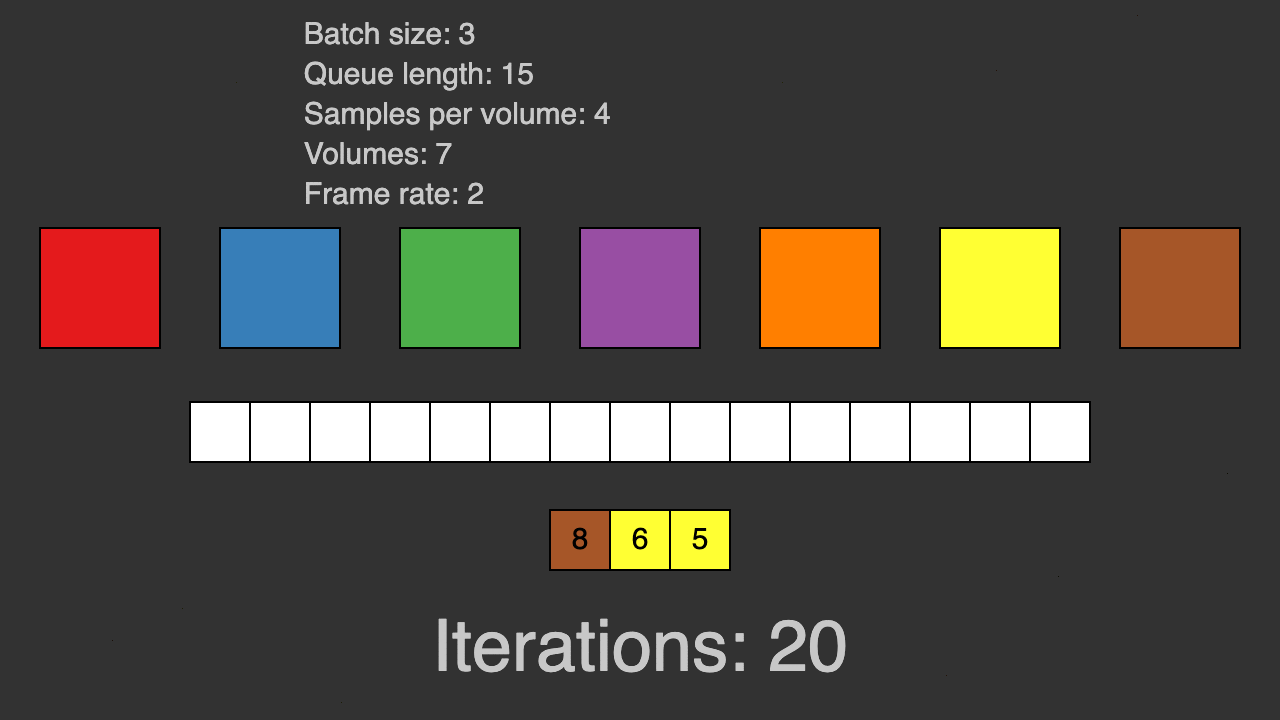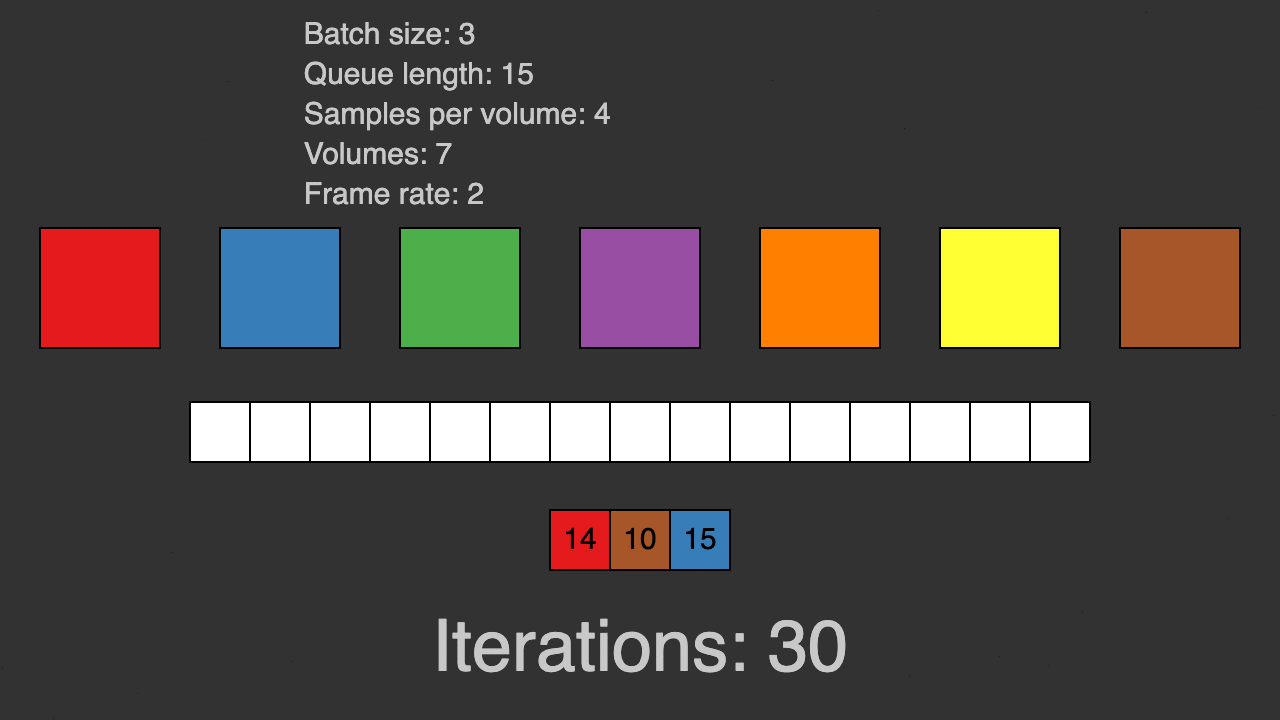
(Queue for patch-based training)
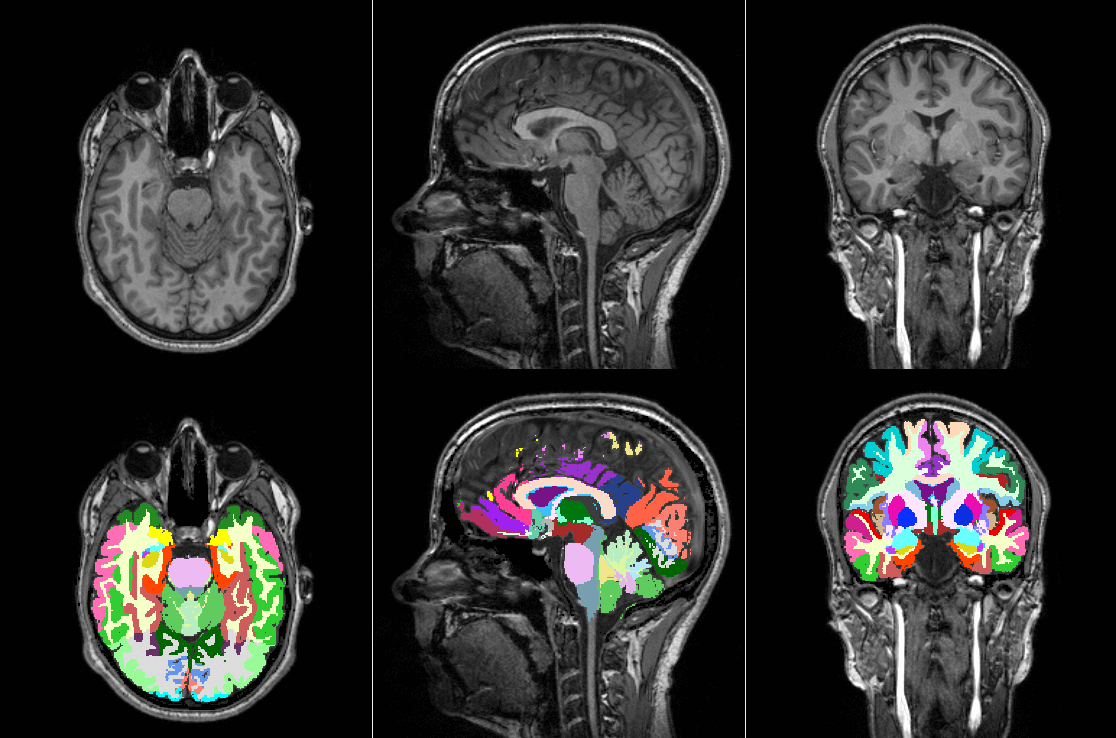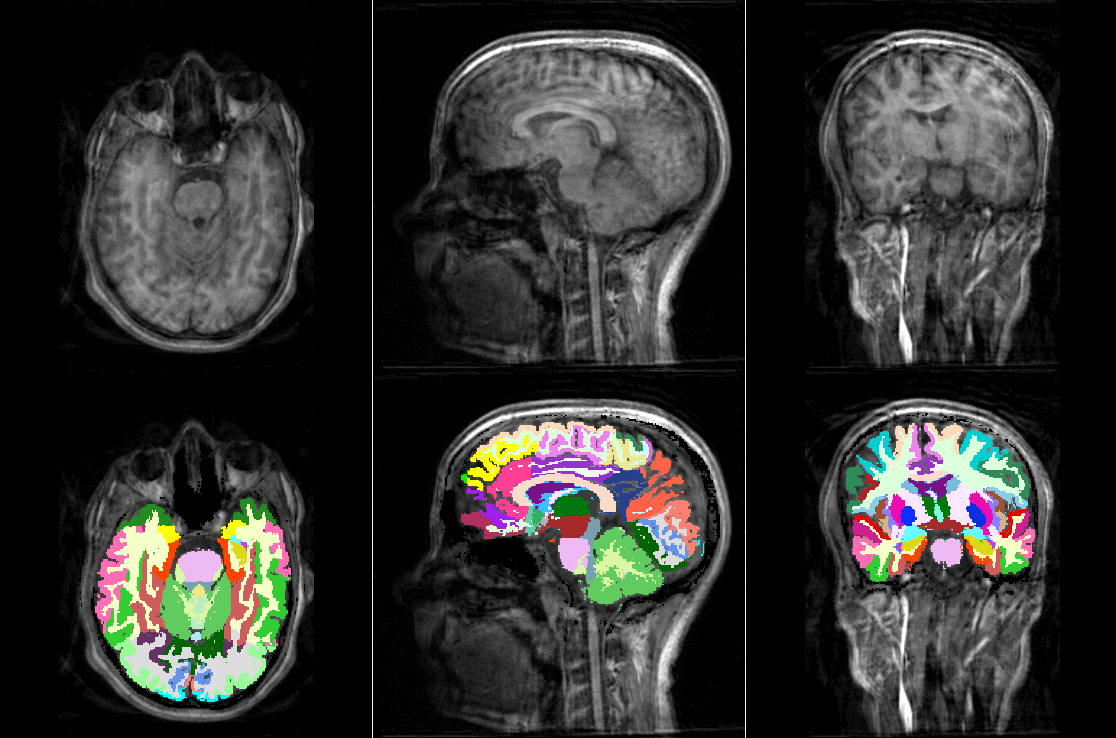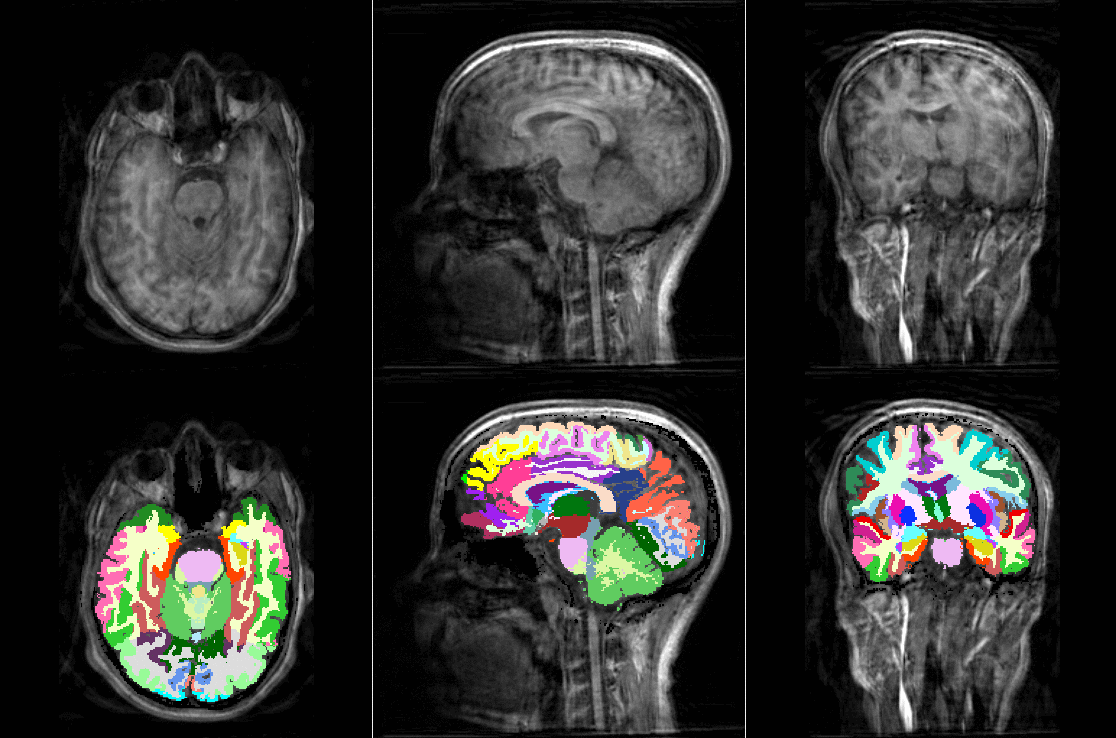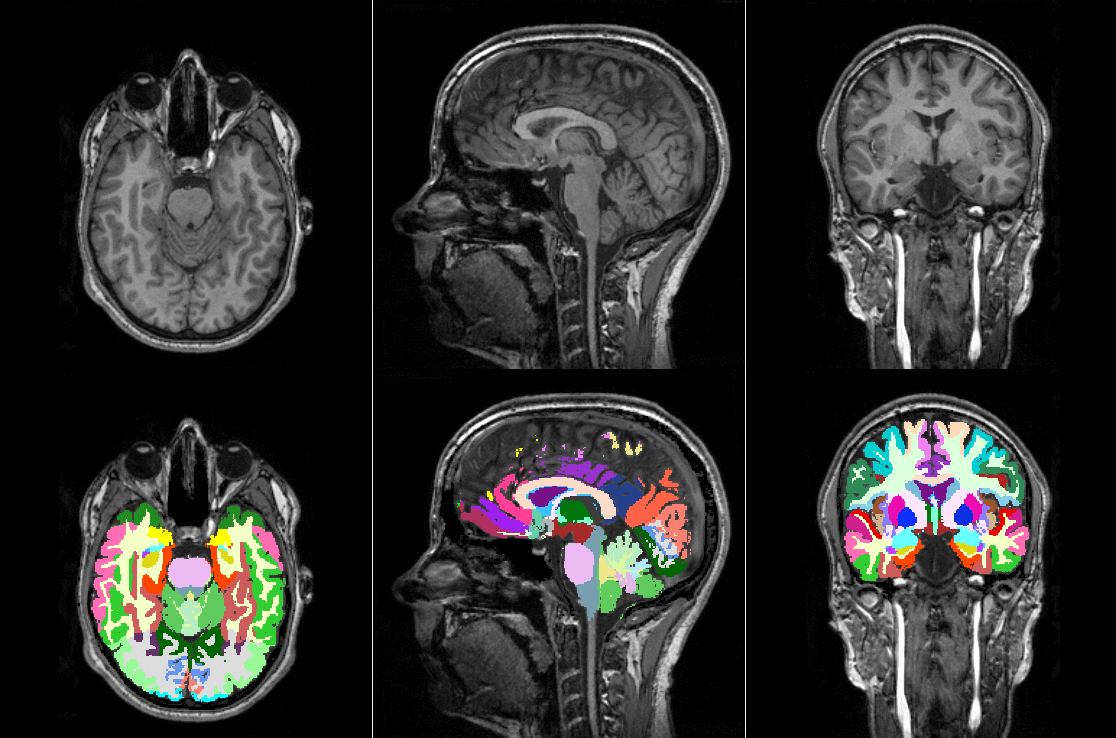
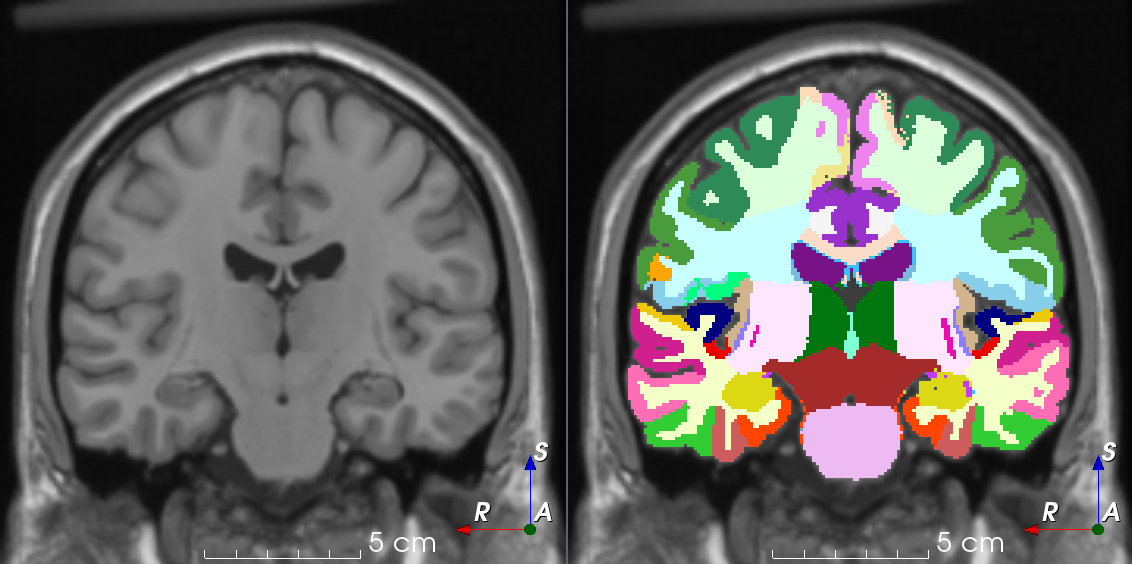
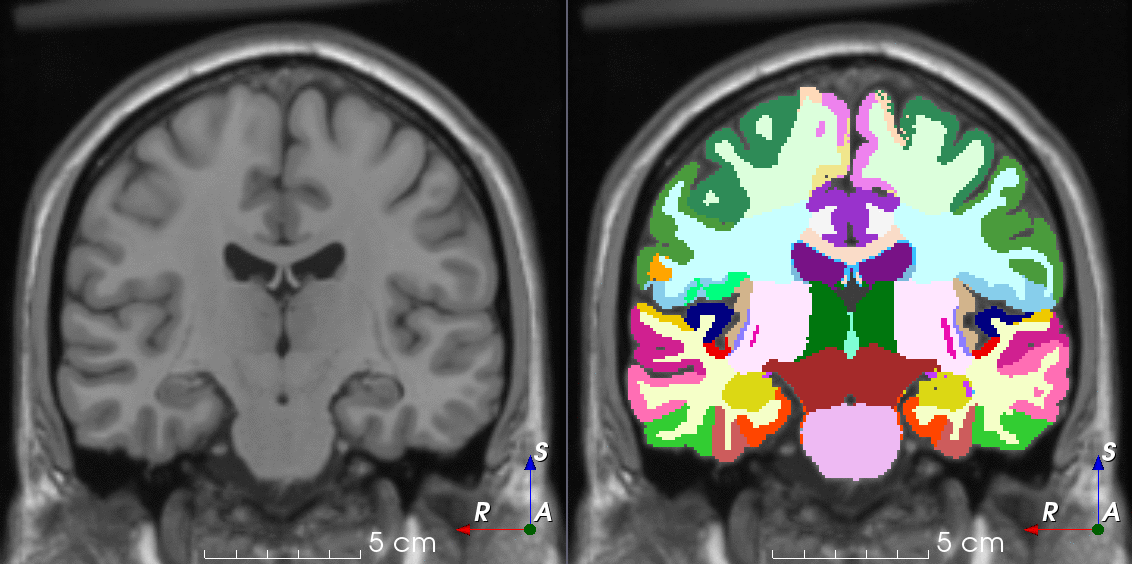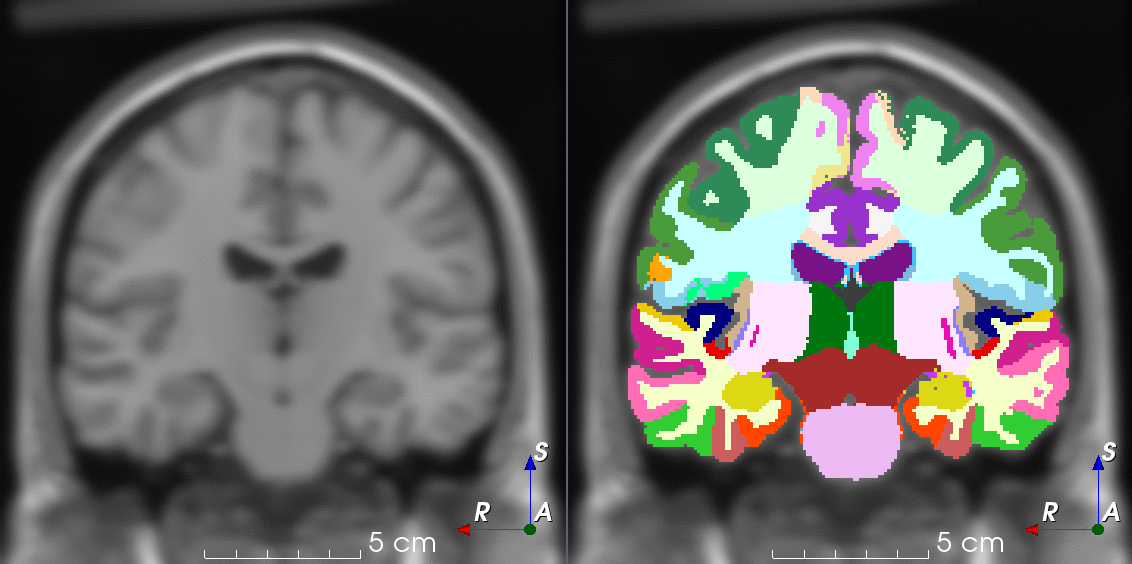
TorchIO is a Python package containing a set of tools to efficiently read, preprocess, sample, augment, and write 3D medical images in deep learning applications written in PyTorch, including intensity and spatial transforms for data augmentation and preprocessing. Transforms include typical computer vision operations such as random affine transformations and also domain-specific ones such as simulation of intensity artifacts due to MRI magnetic field inhomogeneity or k-space motion artifacts.
This package has been greatly inspired by NiftyNet, which is not actively maintained anymore.
Credits
If you like this repository, please click on Star!
If you use this package for your research, please cite our paper:
BibTeX entry:
@article{perez-garcia_torchio_2021,
title = {TorchIO: a Python library for efficient loading, preprocessing, augmentation and patch-based sampling of medical images in deep learning},
journal = {Computer Methods and Programs in Biomedicine},
pages = {106236},
year = {2021},
issn = {0169-2607},
doi = {https://doi.org/10.1016/j.cmpb.2021.106236},
url = {https://www.sciencedirect.com/science/article/pii/S0169260721003102},
author = {P{\'e}rez-Garc{\'i}a, Fernando and Sparks, Rachel and Ourselin, S{\'e}bastien},
}
This project is supported by the following institutions:
- Engineering and Physical Sciences Research Council (EPSRC) & UK Research and Innovation (UKRI)
- EPSRC Centre for Doctoral Training in Intelligent, Integrated Imaging In Healthcare (i4health) (University College London)
- Wellcome / EPSRC Centre for Interventional and Surgical Sciences (WEISS) (University College London)
- School of Biomedical Engineering & Imaging Sciences (BMEIS) (King's College London)
Getting started
See Getting started for installation instructions and a Hello, World! example.
Longer usage examples can be found in the tutorials.
All the documentation is hosted on Read the Docs.
Please open a new issue if you think something is missing.
Contributors
Thanks goes to all these people (emoji key):
This project follows the all-contributors specification. Contributions of any kind welcome!
History
0.18.0 (29-11-2020)
- Add
FPGdataset - Optimize cropping in samplers
- Optimize implementation of
UniformSamplerandWeightedSampler - Fix non integer labels in Colin 27 version 2008 (#223)
- Add
RandomAnisotropytransform - Add
RandomGammatransform - Add
ScalarImageandLabelMapclasses - Add
RandomLabelsToImagetransform - Add support for more input types in
Transform - Add support for 4D images
- Add ICBM dataset
- Add support to specify axes as anatomical labels
- Add
SpatialTransformandIntensityTransform - Replace
ImagesDatasetwithSubjectsDataset - Add support to pass multiple paths to
Image - Refactor reproducibility features
- Add deterministic versions of all random transforms
- Add support to invert transforms
- Add support for SimpleITK > 1
- Add
pre-committo help contributions and improve coda quality - Add DICOM data for testing
- Add some visualization support (
Subject.plot,Image.plot) - Add support to pass parameters per axis, e.g. for
RandomAffine(#346) - Remove deprecated transforms
ResampleandCenterCropOrPad
0.17.0 (23-06-2020)
- Add transforms history to
Subjectattributes to improve traceability - Add support to use an initial transformation in
Resample - Add support to use an image file as target in
Resample - Add
meanargument toRandomNoise - Add tensor support for transforms
- Add support to use strings as interpolation argument
- Add support for 2D images
- Add attribute access to
SubjectandImage - Add MNI and 3D Slicer datasets
- Add
intensityargument toRandomGhosting - Add
translationargument toRandomAffine - Add shape, spacing and orientation attributes to
ImageandSubject - Refactor samplers
- Refactor inference classes
- Add 3D Slicer extension
- Add ITK-SNAP datasets
- Add support to take NumPy arrays as transforms input
- Optimize cropping using PyTorch
- Optimizing transforms by reducing number of tensor copying
- Improve representation (
repr()) ofImage - Use lazy loading in
Image
0.16.0 (21-04-2020)
- Add advanced padding options for
RandomAffine - Add reference space options in
Resample - Add probability argument to all transforms
- Add
OneOfandComposetransforms to improve composability
0.15.0 (07-04-2020)
- Refactor
RandomElasticDeformationtransform - Make
Subjectinherit fromdict
0.14.0 (31-03-2020)
- Add
datasetsmodule - Add support for DICOM files
- Add documentation
- Add
CropOrPadtransform
0.13.0 (24-02-2020)
- Add
Subjectclass - Add random blur transform
- Add lambda transform
- Add random patches swapping transform
- Add MRI k-space ghosting artefact augmentation
0.12.0 (21-01-2020)
- Add ToCanonical transform
- Add CenterCropOrPad transform
0.11.0 (15-01-2020)
- Add Resample transform
0.10.0 (15-01-2020)
- Add Pad transform
- Add Crop transform
0.9.0 (14-01-2020)
- Add CLI tool to transform an image from file
0.8.0 (11-01-2020)
- Add Image class
0.7.0 (02-01-2020)
- Make transforms use PyTorch tensors consistently
0.6.0 (02-01-2020)
- Add support for NRRD
0.5.0 (01-01-2020)
- Add bias field transform
0.4.0 (29-12-2019)
- Add MRI k-space motion artefact augmentation
0.3.0 (21-12-2019)
- Add Rescale transform
- Add support for multimodal data and missing modalities
0.2.0 (2019-12-06)
- First release on PyPI.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file torchio-0.18.63.tar.gz.
File metadata
- Download URL: torchio-0.18.63.tar.gz
- Upload date:
- Size: 40.2 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.5.0 importlib_metadata/4.8.1 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.9.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 148e80a996726c59df545d9e1831ce3bae4346403b4cd54a0beb56db037ac4bf |
|
| MD5 | 70df6fdc640cfa8f0ca4d4965c77de7b |
|
| BLAKE2b-256 | ca60504771f2b6b2c72d083cc52fbd2418bbb912e501589ca195d4bbb2300364 |
File details
Details for the file torchio-0.18.63-py2.py3-none-any.whl.
File metadata
- Download URL: torchio-0.18.63-py2.py3-none-any.whl
- Upload date:
- Size: 161.9 kB
- Tags: Python 2, Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/3.5.0 importlib_metadata/4.8.1 pkginfo/1.7.1 requests/2.26.0 requests-toolbelt/0.9.1 tqdm/4.62.3 CPython/3.9.7
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 64200eaa0adf85efdef0af7ddef24de80011bd957e4f663a42480e0c35221284 |
|
| MD5 | 8d3636d3cf4114db5e396b05e9c6eb33 |
|
| BLAKE2b-256 | 320338ac0d5baa043a8db7043ecebb5571216ee4b6f4b5e5cd8a1403c33d24a8 |