computer vision for image-based phenotyping of single cells
Project description
VisCy
VisCy (abbreviation of vision and cyto) is a deep learning pipeline for training and deploying computer vision models for image-based phenotyping at single-cell resolution.
This repository provides a pipeline for the following.
- Image translation
- Robust virtual staining of landmark organelles
- Image classification
- Supervised learning of of cell state (e.g. state of infection)
- Image representation learning
- Self-supervised learning of the cell state and organelle phenotypes
Note:
VisCy has been extensively tested for the image translation task. The code for other tasks is under active development. Frequent breaking changes are expected in the main branch as we unify the codebase for above tasks. If you are looking for a well-tested version for virtual staining, please use release0.2.1from PyPI.
Virtual staining
Demos
-
Virtual staining exercise: Notebook illustrating how to use VisCy to train, predict and evaluate the VSCyto2D model. This notebook was developed for the DL@MBL2024 course and uses UNeXt2 architecture.
-
Image translation demo: Fluorescence images can be predicted from label-free images. Can we predict label-free image from fluorescence? Find out using this notebook.
-
Training Virtual Staining Models via CLI: Instructions for how to train and run inference on ViSCy's virtual staining models (VSCyto3D, VSCyto2D and VSNeuromast).
Gallery
Below are some examples of virtually stained images (click to play videos). See the full gallery here.
| VSCyto3D | VSNeuromast | VSCyto2D |
|---|---|---|
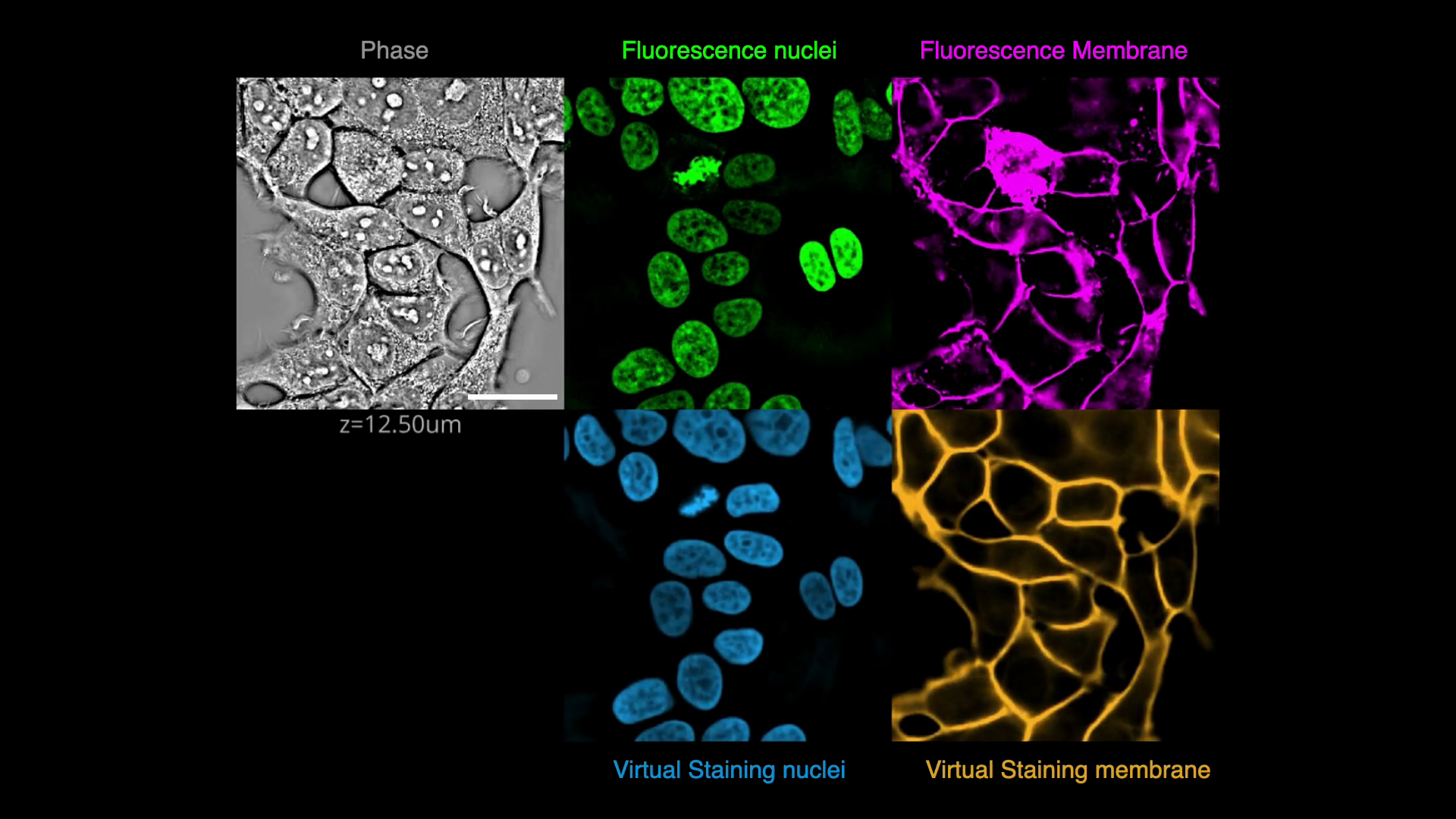 |
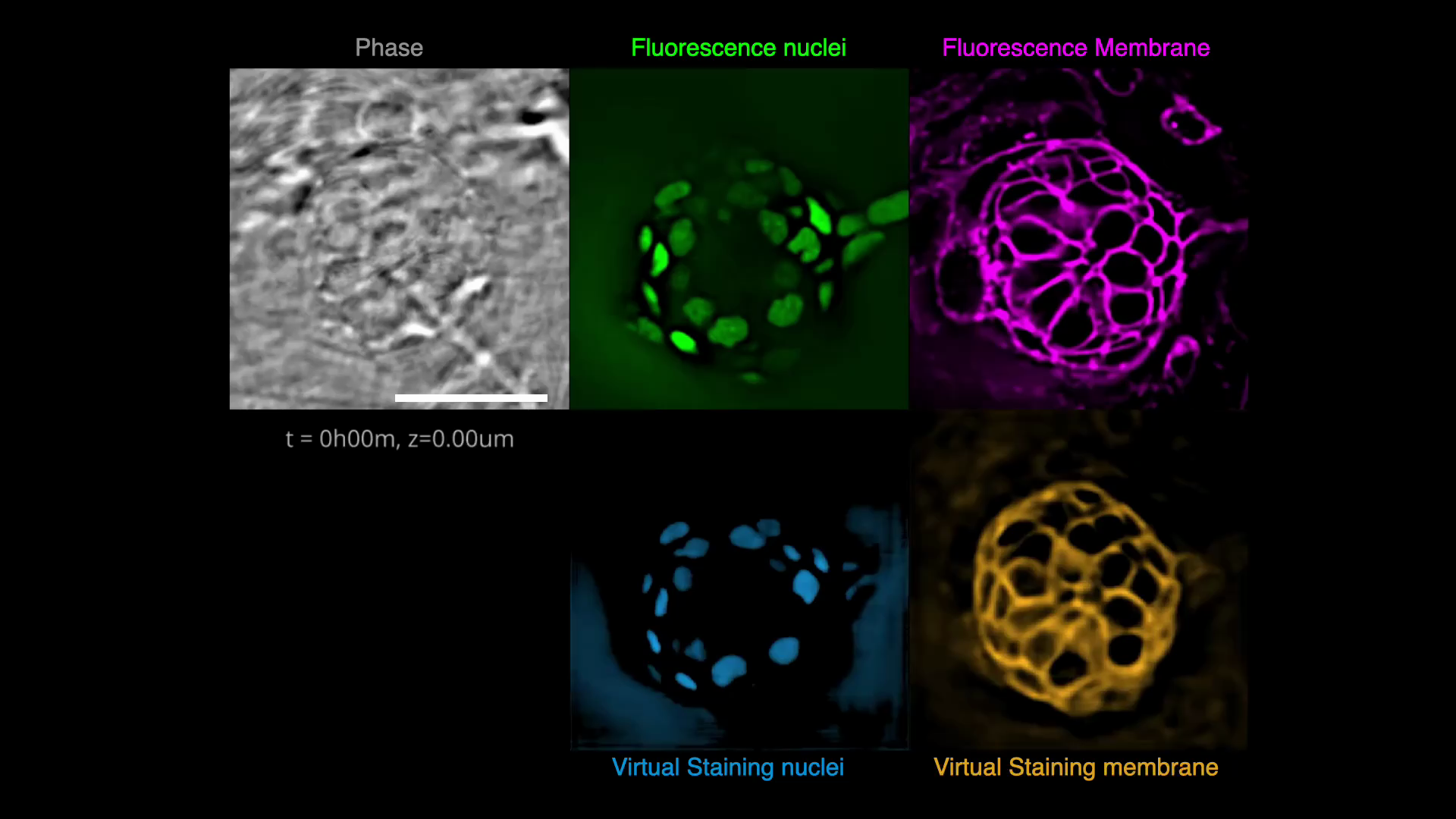 |
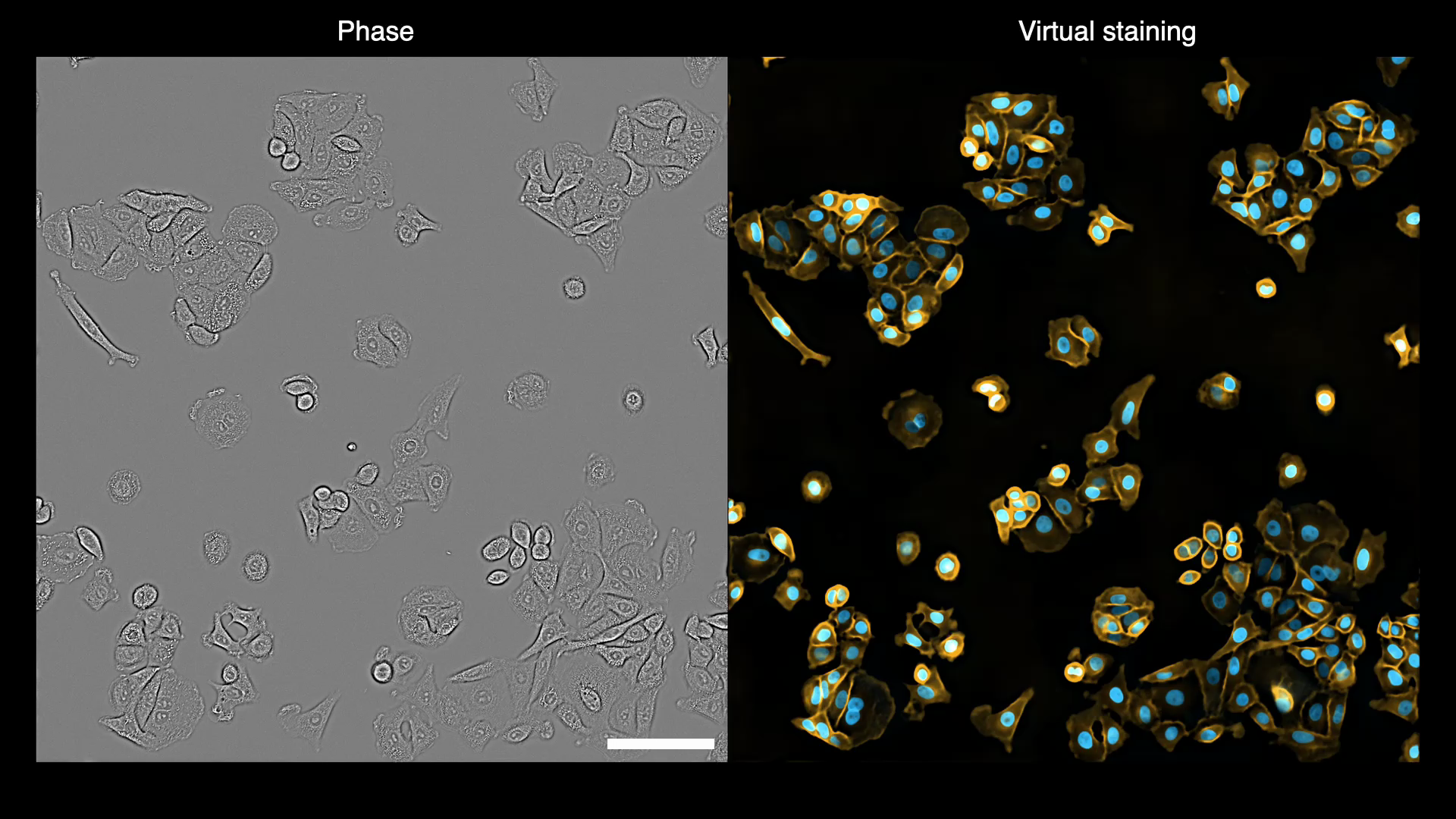 |
Reference
The virtual staining models and training protocols are reported in our recent preprint on robust virtual staining.
This package evolved from the TensorFlow version of virtual staining pipeline, which we reported in this paper in 2020.
Liu, Hirata-Miyasaki et al., 2024
@article {Liu2024.05.31.596901,
author = {Liu, Ziwen and Hirata-Miyasaki, Eduardo and Pradeep, Soorya and Rahm, Johanna and Foley, Christian and Chandler, Talon and Ivanov, Ivan and Woosley, Hunter and Lao, Tiger and Balasubramanian, Akilandeswari and Liu, Chad and Leonetti, Manu and Arias, Carolina and Jacobo, Adrian and Mehta, Shalin B.},
title = {Robust virtual staining of landmark organelles},
elocation-id = {2024.05.31.596901},
year = {2024},
doi = {10.1101/2024.05.31.596901},
publisher = {Cold Spring Harbor Laboratory},
URL = {https://www.biorxiv.org/content/early/2024/06/03/2024.05.31.596901},
eprint = {https://www.biorxiv.org/content/early/2024/06/03/2024.05.31.596901.full.pdf},
journal = {bioRxiv}
}
Guo, Yeh, Folkesson et al., 2020
@article {10.7554/eLife.55502,
article_type = {journal},
title = {Revealing architectural order with quantitative label-free imaging and deep learning},
author = {Guo, Syuan-Ming and Yeh, Li-Hao and Folkesson, Jenny and Ivanov, Ivan E and Krishnan, Anitha P and Keefe, Matthew G and Hashemi, Ezzat and Shin, David and Chhun, Bryant B and Cho, Nathan H and Leonetti, Manuel D and Han, May H and Nowakowski, Tomasz J and Mehta, Shalin B},
editor = {Forstmann, Birte and Malhotra, Vivek and Van Valen, David},
volume = 9,
year = 2020,
month = {jul},
pub_date = {2020-07-27},
pages = {e55502},
citation = {eLife 2020;9:e55502},
doi = {10.7554/eLife.55502},
url = {https://doi.org/10.7554/eLife.55502},
keywords = {label-free imaging, inverse algorithms, deep learning, human tissue, polarization, phase},
journal = {eLife},
issn = {2050-084X},
publisher = {eLife Sciences Publications, Ltd},
}
Library of virtual staining (VS) models
The robust virtual staining models (i.e VSCyto2D, VSCyto3D, VSNeuromast), and fine-tuned models can be found here
Pipeline
A full illustration of the virtual staining pipeline can be found here.
Installation
-
We recommend using a new Conda/virtual environment.
conda create --name viscy python=3.10 # OR specify a custom path since the dependencies are large: # conda create --prefix /path/to/conda/envs/viscy python=3.10
-
Install a released version of VisCy from PyPI:
pip install viscy
If evaluating virtually stained images for segmentation tasks, install additional dependencies:
pip install "viscy[metrics]"
Visualizing the model architecture requires
visualdependencies:pip install "viscy[visual]"
-
Verify installation by accessing the CLI help message:
viscy --help
For development installation, see the contributing guide.
Additional Notes
The pipeline is built using the PyTorch Lightning framework. The iohub library is used for reading and writing data in OME-Zarr format.
The full functionality is tested on Linux x86_64 with NVIDIA Ampere GPUs (CUDA 12.4).
Some features (e.g. mixed precision and distributed training) may not be available with other setups,
see PyTorch documentation for details.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file viscy-0.2.1.tar.gz.
File metadata
- Download URL: viscy-0.2.1.tar.gz
- Upload date:
- Size: 17.7 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.1 CPython/3.10.14
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | eb422122260ab99014081ecef3d880f596e3bc1886624c2d56c9160613ef4e8a |
|
| MD5 | 2e71bc4d8051b8e9e3a08ce80be4367f |
|
| BLAKE2b-256 | f2a8faf69e8454faaa3857c96ac8a08ad836fb13ed0f6b23f1bd7f7c51f1bed6 |
File details
Details for the file viscy-0.2.1-py3-none-any.whl.
File metadata
- Download URL: viscy-0.2.1-py3-none-any.whl
- Upload date:
- Size: 103.4 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.1 CPython/3.10.14
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 56a33be450b022cbdf16357ae237c6084d7d0d02a2dd50585c5e68ff3f940643 |
|
| MD5 | ae40be484a1379793bc29b95cb5f11b3 |
|
| BLAKE2b-256 | 3a3bac436140783522349d4774d120cc96e8b4051c293feb0c0ebe353d9b850b |












