napari plugin for nd tiff folders with OpenCl deskew
Project description
napari-ndtiffs
napari plugin for nd tiff folders with optional CUDA or OpenCL-based deskewing.
Built-in support for folders of (skewed) lattice light sheet tiffs.
This napari plugin was generated with Cookiecutter using with @napari's cookiecutter-napari-plugin template.
Features
- Drag and drop a folder of tiffs onto napari window to view easily
- (currently designed to detect lattice light sheet tiffs, but easily adjustable)
- If lattice
Settings.txtfile is found, will deskew automatically (only if necessary) - Lazily loads dataset on demand. quickly load preview your data.
- Handles
.ziparchives as well! Just directly compress your tiff folder, then drop it into napari. - All OpenCL deskewing, works on GPU as well as CPU, falls back to scipy if PyOpenCL is unavailable.
- CuPy-based deskewing will work for cards with NVIDIA GPUs that support CUDA. CuPy 8.x releases should work, although CuPy >= 9 is recommended. If CuPy is unavailable, the PyOpenCL implementation is used instead.
It would not be hard to support arbitrary filenaming patterns! If you have a folder of tiffs with a consistent naming scheme and would like to take advantage of this plugin, feel free to open an issue!
Installation
You can install napari-ndtiffs via pip:
pip install napari-ndtiffs
To also install PyOpenCL (for faster deskewing):
pip install napari-ndtiffs[opencl]
On NVIDIA GPUs with CUDA support, the CuPy implementation may be faster than PyOpenCL. CuPy also has experimental support for AMD GPUs via HIP/ROCm. See the CuPy installation instructions
Usage
In most cases, just drop your folder onto napari, or use viewer.open("path")
Overriding parameters
You can control things like voxel size and deskewing angle as follows:
from napari_ndtiffs import parameter_override
import napari
viewer = napari.Viewer()
with parameter_override(angle=45, name="my image"):
viewer.open("path/to/folder", plugin="ndtiffs")
Valid keys for parameter_override include:
- dx: (
float) the pixel size, in microns - dz: (
float)the z step size, in microns - deskew: (
bool) whether or not to deskew, (by default, will deskew if angle > 0, or if a lattice metadata file is detected that requires deskewing) - angle: (
float) the angle of the light sheet relative to the coverslip - padval: (
float) the value with which to pad the image edges when deskewing (default is 0) - contrast_limits: (
2-tuple of int) (min, max) contrast_limits to use when viewing the image - name: (
str) an optional name for the image
Sample data
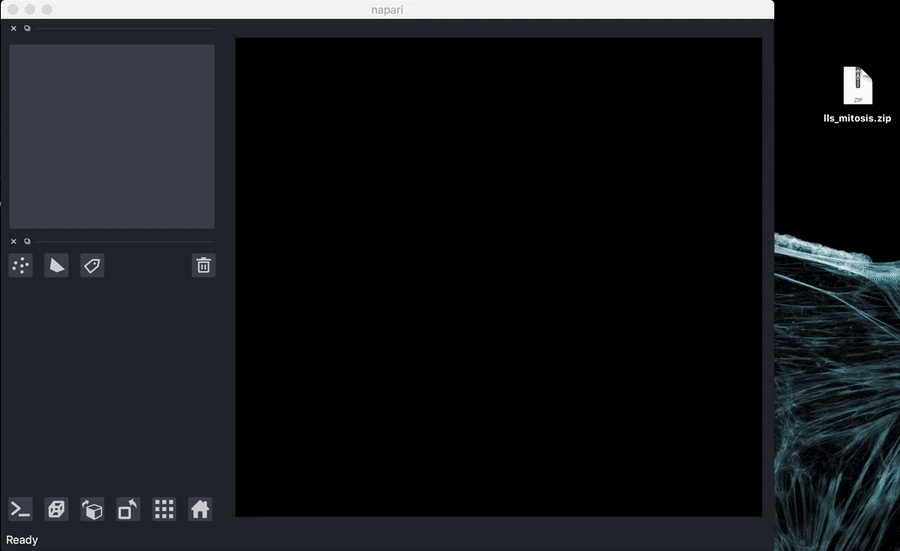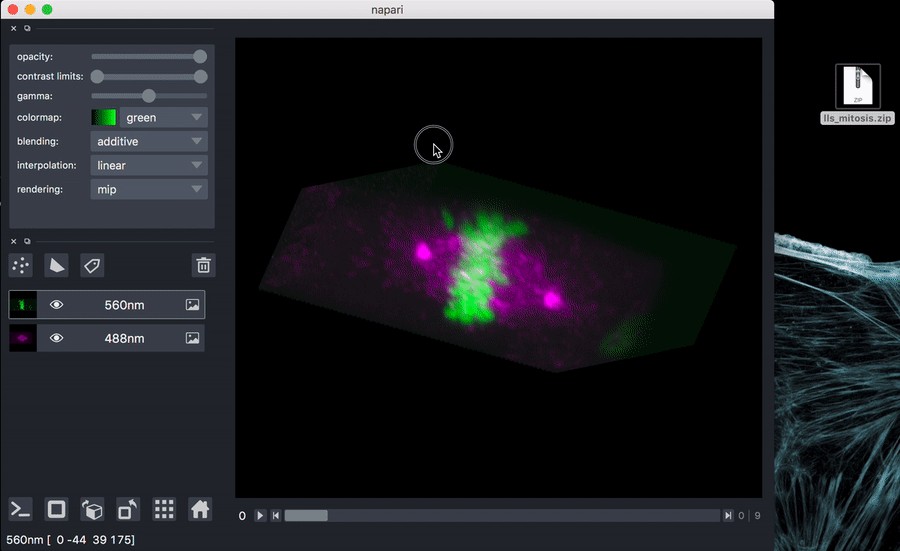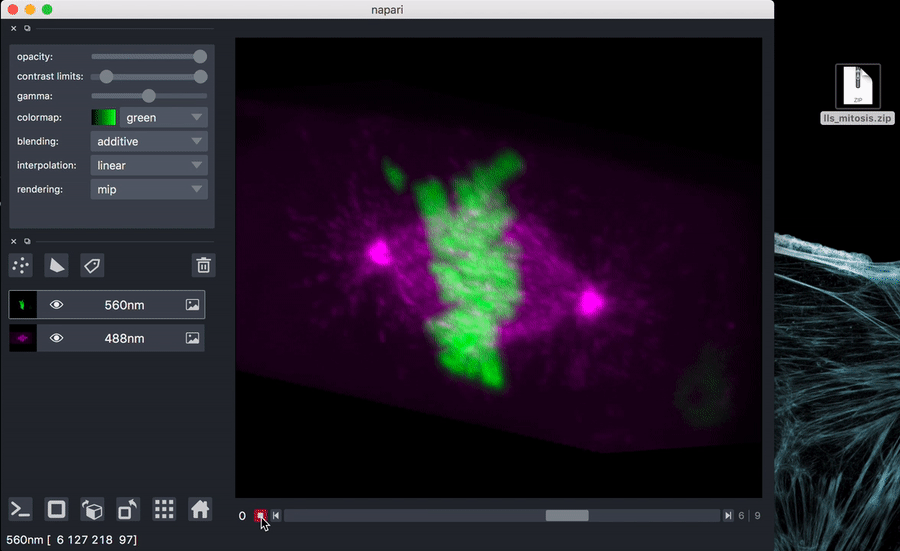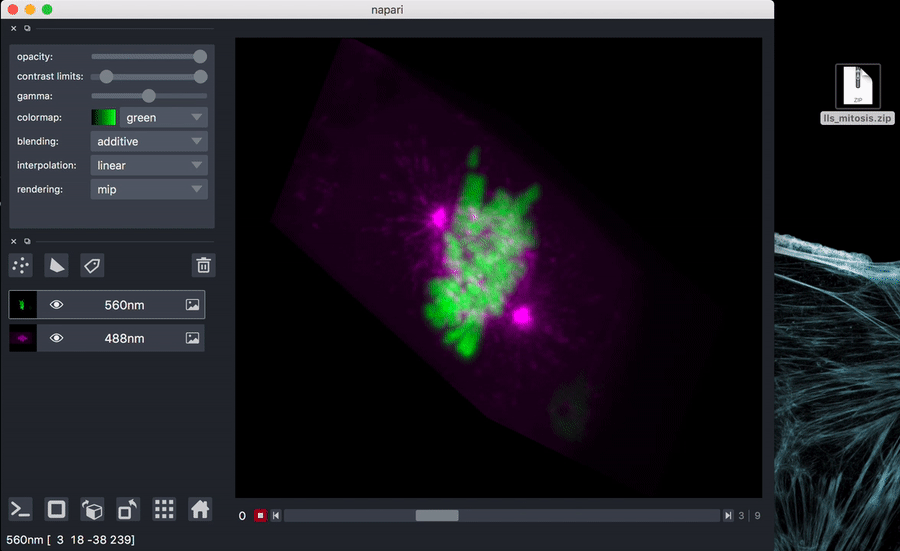
Try it out with test data: download sample data
You can unzip if you like, or just drag the zip file onto the napari window.
Or, from command line, use:
napari path/to/lls_mitosis.zip
Debugging
To monitor file io and deskew activity, enter the following in the napari console:
import logging
logging.getLogger('napari_ndtiffs').setLevel('DEBUG')
Contributing
Contributions are very welcome. Tests can be run with tox, please ensure the coverage at least stays the same before you submit a pull request.
License
Distributed under the terms of the BSD-3 license, "napari-ndtiffs" is free and open source software
Issues
If you encounter any problems, please file an issue along with a detailed description.
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
Built Distribution
File details
Details for the file napari_ndtiffs-0.2.1.tar.gz.
File metadata
- Download URL: napari_ndtiffs-0.2.1.tar.gz
- Upload date:
- Size: 9.1 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.1 CPython/3.11.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 4e290f2d87e4a9a1020ff5b8ad66f4a6d92f5ea6daadd60b937beae4b5312054 |
|
| MD5 | dceb53d80fb29fc01f1b4586f2c8a98e |
|
| BLAKE2b-256 | 5dc3eaabb32578a5e15fd85b867fbd21fc5d120b7ba7c413973fcdb1ac11b8e1 |
File details
Details for the file napari_ndtiffs-0.2.1-py3-none-any.whl.
File metadata
- Download URL: napari_ndtiffs-0.2.1-py3-none-any.whl
- Upload date:
- Size: 17.6 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/4.0.1 CPython/3.11.2
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 41dc55610a8a985faace3d11f8d9e3af5cbd9aa864f46dae3e3bcd0b2773d929 |
|
| MD5 | 5ea39b086bf32a3e8bfc5727f636b6d7 |
|
| BLAKE2b-256 | 80f81311b720c4879477aba839abe6c09c010796f853de56762e2edc26e71e2c |