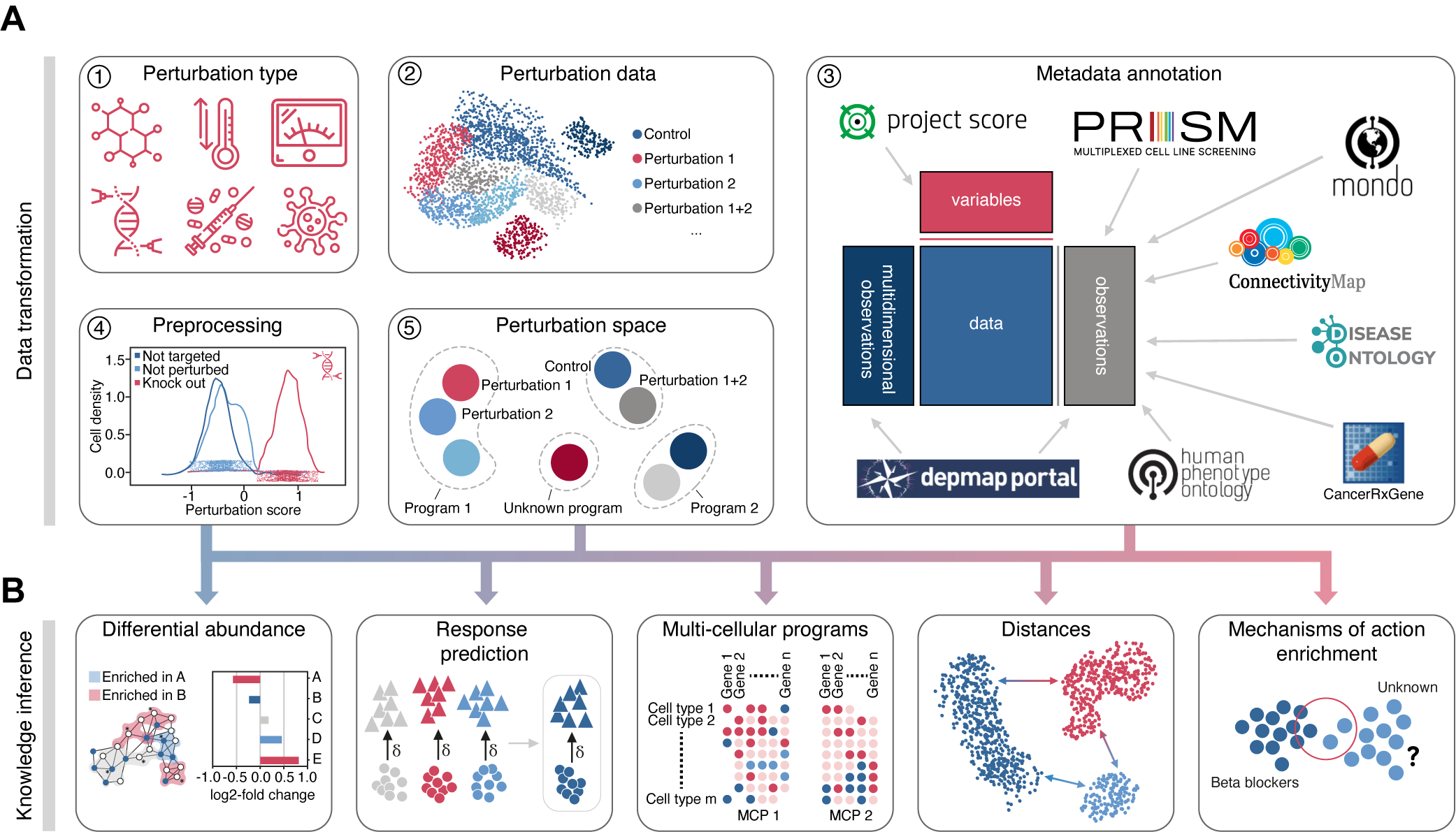
Perturbation Analysis in the scverse ecosystem.
Project description
pertpy
Documentation
Please read the documentation.
Installation
We recommend installing and running pertpy on a recent version of Linux (e.g. Ubuntu 24.04 LTS). No particular hardware beyond a standard laptop is required.
You can install pertpy in less than a minute via pip from PyPI:
pip install pertpy
if you want to use scCODA or tascCODA, please install pertpy as follows:
pip install pertpy[coda]
If you want to use the differential gene expression interface, please install pertpy by running:
pip install pertpy[de]
Citation
Project details
Release history Release notifications | RSS feed
Download files
Download the file for your platform. If you're not sure which to choose, learn more about installing packages.
Source Distribution
pertpy-0.9.1.tar.gz
(3.0 MB
view details)
Built Distribution
pertpy-0.9.1-py3-none-any.whl
(209.0 kB
view details)
File details
Details for the file pertpy-0.9.1.tar.gz.
File metadata
- Download URL: pertpy-0.9.1.tar.gz
- Upload date:
- Size: 3.0 MB
- Tags: Source
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.12.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 0714ea9385458519c10f5ae4aff3ddab4a299eb9d1b0d5512eb9c531f1c150cb |
|
| MD5 | 334a94f4febf095c2736a733fbe3e6f6 |
|
| BLAKE2b-256 | 275b2af9752b1c32cbd453f38c111bc03f8b2b9ab8650b2412b1117ceaba6b16 |
Provenance
File details
Details for the file pertpy-0.9.1-py3-none-any.whl.
File metadata
- Download URL: pertpy-0.9.1-py3-none-any.whl
- Upload date:
- Size: 209.0 kB
- Tags: Python 3
- Uploaded using Trusted Publishing? No
- Uploaded via: twine/5.1.0 CPython/3.12.5
File hashes
| Algorithm | Hash digest | |
|---|---|---|
| SHA256 | 3525b0d83878a23463e920cb533fd55f42db7d326cfec60e0445ec200ffdfdbd |
|
| MD5 | a4e344443ea0cd6ebb8ecf8a671d140f |
|
| BLAKE2b-256 | 797e4fae38f064205ef9d1b064328b4dfedf070f2164b8a7996cd47cfd7b096a |